Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Andrew Ford, Charmaine Brown and Chen-Hsiung Yeh*
Received: June 27, 2017; Published: July 03, 2017
Corresponding author: Chen-Hsiung Yeh, Circulogene Theranostics, 3125 Independence Dr, Suite 301, Birmingham, AL 35209, USA
DOI: 10.26717/BJSTR.2017.01.000166
Keywords: Heterochromatin; Euchromatin; Cell-free DNA; Liquid biopsy; Next-generation sequencing
Abbreviations: cfDNA: cell-free DNA; NGS: Next-generation Sequencing
The human genome contains ~30,000 genes that need to be expressed in specific cells at precise times. The DNA in the nucleus exists in two forms that reflect the level of activity of the cell. Through electronic microscopy, heterochromatin appears as small, darkly staining, irregular particles scattered throughout the nucleus or accumulated adjacent to the nuclear envelope; on the other hand, euchromatin is dispersed and not readily stainable. Euchromatin comprises the most active portion of the genome within the cell nucleus and is enriched in genes, and is often under active transcription, whereas non-coding sequences are most abundant in heterochromatin. The position in the chromosome and/or changes to the chromatin structure can significantly influence gene expression: genes are usually silenced when the chromatin is condensed, and are active when chromatin is open. These dynamic chromatin states can be regulated at multiple levels via reversible epigenetic patterns of DNA methylation, histone or nucleosome modifications as well as tissue-specific transcriptional activators and repressors [1].
Circulating cell-free DNA (cfDNA) has been recently established as a diagnostic, predictive and prognostic biomarker [2]. In healthy individuals, cfDNA fractions are mostly derived from apoptosis of various normal cells, whereas cfDNA of cancer patients represents a mixture of apoptosis, necrosis, autophagy, or mitotic catastrophe [3]. Necrosis produces relatively long fragments of DNA, about 10 kb in length, while in apoptosis, the activation of endogenous endonucleases lead to the cleavage of chromatin DNA into small inter- nucleosomal fragments of ~170 bp [4]. Consequently, chromatin structure-dependent fragmentation of cfDNA has important implications for the analysis and application of cfDNA liquid biopsy in clinical testing.
Figure 1:TP53 tissue expression correlated inversely with cfDNA sequencing coverage.
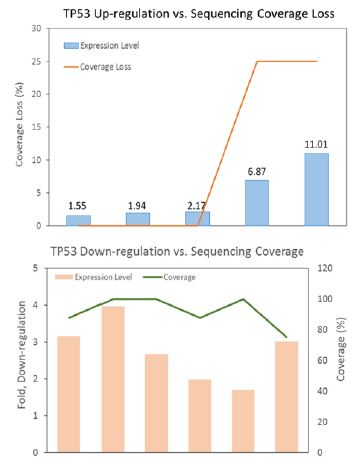
It is in agreement that the cfDNA retains characteristics previously noted in genome-wide analysis of chromatin structure, and the fragment size distribution and the read spacing revealed that the nucleosome-guided fragmentation of cfDNA is not random, reflecting a general picture of gene expression [5,6], and may further indicate an unbalanced representation of genomic sequences. Since biased fragmentation patterns of cfDNA could affect the uniformity of genome coverage analyzed by next-generation sequencing (NGS), we examined the relationship between cfDNA depth of coverage and tumor tissue RNA expression level of TP53 gene from different ovarian cancer patients. TP53 cfDNA sequencing displayed lower coverage depth when tissue TP53 expression was up-regulated, while down-regulation of TP53 tissue expression was concomitant with higher cfDNA sequencing coverage (Figure 1), consistent with the idea that actively transcribed genes (chromosome regions devoid of histones/nucleosomes binding) are structurally more open and accessible to nuclease attack when releasing into bloodstream. This observation highly resembles what was observed in WGS data of randomly selected samples from the 1000 Genome Project [7].
Our findings that gained or lost NGS coverage depth in cfDNA negatively correlated with transcriptional activity in tissue provided the first hand information of interplay between tissue RNA level and blood cfDNA fragmentation at single-gene resolution. The association of cfDNA fragment copy number with the expression levels in respective locus may aid in detection of various pathologies, including the presence of different types of neoplasms. Mapping and mining cfDNA fragment may also help in the development of novel biomarkers reflecting pathological changes in chromatin architecture. Technically and clinically, reproducible fragmentation patterns of cfDNA could be used to harness higher sensitivity and accuracy in NGS detection of certain gene mutations based on their chromatin positions.


