Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Tsann-Long Hwang* and Chih-Hong Lo
Received: January 07, 2019; Published: January 22, 2019
*Corresponding author: Tsann-Long Hwang, Department of Surgery, Chang Gung Memorial Hospital, 5, Fu-Hsin Street, Kwi-San, Tau-Yuan County, Taiwan
DOI: 10.26717/BJSTR.2019.13.002392
Autophagy was found to effect on cancer initiation and progression, due to the overlapping of autophagic and cancer signaling pathways. Pancreatic ductal cancer had elevated autophagy under the basal conditions than those of other epithelial cancers. Cancer stem cells (CSCs) were found not only a model of carcinogenesis, but also related to the mechanism of drug resistance and tumor recurrence in recent years. The role of autophagy and related genes in pancreatic cancer stem cells for drug resistance of pancreatic cancer was investigated. The expressions of autophagy of CD44+/- in pancreatic cell lines were investigated, and the expressions of autophagy regulatory genes were also studied. Our results showed that LC3‑II proteins had significant expression in the BxPC-3 and Capan-1 pancreatic cancer cells.
The percentage of viable cells for CD44+ stem cells was higher autophagy after gemcitabine was added in both BxPC-3 and Capan-1 cells. The difference could demonstrate that resistance of gemcitabine treatment in CD44+ stem cells related to its higher autophagy activity. RQ vs both pancreatic cancer cell lines treated with gemcitabine 50 nM 24 hours showed significant expression of ATG12, UVRAG, MAP1LC3B, and MAP1LC3A genes showed significant expression. The difference of MAP1LC3B, and M. AP1LC3A genes was also demonstrated between CD44+ and CD44- . We concluded that resistance of gemcitabine treatment in CD44+ stem cells for pancreatic ductal cancer was related to its higher autophagy activity. Blockage of ATG12, UVRAG, MAP1LC3B, and MAP1LC3A cancer stem cells related genes might contribute to the effectiveness of gemcitabine chemotherapy for patients with pancreatic ductal cancer.
Keywords: Pancreatic Ductal Cancer; Autophagy; Cancer Stem Cell; Drug Resistance
Pancreatic ductal adenocarcinoma (PDAC) is the fourth leading cause of cancer death in United States and is ranked as the 10th~11th cancer frequently associated the cancer mortality in Taiwan. In Taiwan, the overall 5-year survival rate is still around 5 to 10% [1-9] and the 5-year survival rate of the patients with resectable PCDAC is about 15-20% [10]. The genetic or molecular changes of PDAC have been studied for years. Most previous studies of tumor samples have been focused primarily on analysis of the tumor DNA, and more recently on analyses of the tumor mRNA, by aiming at identifying gross genomic alternations, specific genes with mutations, or mRNA expression [11-20]. A regulated mechanism that achieves the removal and reutilization of tumor cells is autophagy. It plays a pro-survival role and can be up-regulated in response to both external and intracellular factors. Autophagy is considered a self-defense mechanism, where macromolecules and complete organelles are engulfed in peri-nuclear double membrane vesicles and degraded in lysosomes. The role of autophagy in cancer depends mainly on the tumor stage. It may have a dual role, as a tumor suppressor in normal cells by degrading onco-proteins, later, allowing cancer cells to survive during metabolic stress [21-24]. Although autophagy is a mechanism of tumor suppression, it confers stress tolerance that enables tumor cells to survive under adverse conditions by recycling of nutrients for metabolic needs, a fundamental aspect of tumor progression.
Autophagy is generated by tumor hypoxia and anaerobic glycolysis, whereas angiogenesis maintains low autophagic activity. It localizes to hypoxic regions of tumors most distal to blood vessels where supports tumor cell survival [25-26]. LC3 is a key component of autophagy, and it has been used as a marker of autophagy. Signaling pathways that promote autophagy are potential candidates for inhibitor development. Pancreatic cancers have more elevated autophagy under basal conditions than those of other epithelial cancers [27]. A clinic-pathological study of 71 archival pancreatic cancer tissues demonstrated that autophagy is activated in pancreatic cancer cells and correlates with poor patient outcome [28]. A stronger LC3 intensity expression level in the peripheral area of the pancreatic cancer tissue by immunohistochemical staining with LC3 antibody was found. Stem cells are unique in their ability to self-renew and differentiate into various cells in the body, which are important in development, tissue renewal and a range of disease processes. Therefore, it is predicted that autophagy would be crucial for the quality control mechanisms and maintenance of cellular homeostasis in various stem cells given their relatively long life in the organisms. In contrast to the extensive body of knowledge available for somatic cells, the role of autophagy in the maintenance and function of stem cells is only beginning to be revealed as a result of recent studies.
Recent studies of different knockout mice models have defined the roles of various autophagy genes and related pathways in the regulation of the maintenance, expansion and differentiation of various stem cells [29-32]. Autophagy is regulated by autophagyrelated genes (ATGs), which play significant roles in autophagosome formation and autophagy regulation. Silencing of some essential ATGs, such as Atg3, Atg4, ATg6/Beclin-1, Atg10, and Atg12, can sensitize cancer cells to a wide spectrum of stressful condition [33]. The autophagicmediators and corresponding proteins can integrate into cancer cell signaling networks and regulate cell survival or death. The understanding of autophagy – cancer relationships can contribute to pancreatic cancer treatment. Modulation of autophagy may provide a possible way to cancer therapy. The following study was designed to investigate the role of autophagy and related genes in the pancreatic stem cells of PDAC and its significance for drug treatment.
CSCs was identified by means of the surface marker CD44, CD24, ESA(Ep-CAM) and CD133. CSCs were isolated by magnetic bead sorting using the MACS system (Miltenyi Biotech). For this purpose, dissociated cells were suspended in PBS. Cells were then incubated with a surface marker monoclonal antibody labeled with Micro Beads (Miltenyi Biotech) for 30 min at 4°C, and the CSCs surface marker positive cells were enriched using a MACS magnet and MS columns (Miltenyi Biotech). All MACS procedures were performed according to the manufacturer’s instructions. The purity of isolated cells was determined by standard flow cytometry analysis using an APC-labeled antibody against human CSCs surface marker. CSCs was isolated by fluorescence activated cell sorting (FACS) for the markers CD44, CD24, ESA and CD133. Dissociated cells were counted and transferred to a 5-ml tube, washed twice with HBSS containing 2% heat-inactivated FBS, and re-suspended in HBSS with 2% FBS at concentration of 106 per 100ul.
Sandoglobin solution (1mg/ml) was then added to the sample at a dilution of 1:20 and the sample was incubated on ice for 20min. The sample was washed twice with HBSS/2% FBS and re-suspended in HBSS/2% FBS. Antibodies were added and incubated for 20min on ice, and the sample was washed twice with HBSS/2% FBS. When needed, a secondary antibody was added by re-suspending the cells in HBSS/2%FBS followed by a 20-min incubation. After another washing, cells were re-suspended in HBSS/2% FBS containing 4’,6-diamidino-2-phenylindole (DAPI;1ug/ml final concentration). The antibodies used were anti-CD44 allophycocyanin(APC), anti- CD24 phycoerythrin(PE), anti–ESA-FITC and anti-CD133 APC, each at a dilution of 1:40. Dead cells were eliminated by using the viability dye DAPI. Flow cytometry was done using a FACSAria (BD Immunocytometry Systems). Side scatter and forward scatter profiles were used to eliminate cell doublets. Cells were routinely sorted twice, and the cells were reanalyzed for purity.
PC cell lines BxPC-3 and PANC-1 and AsPC-1 were purchased from Bioresource Collection and Research Center (Hsinchu, Taiwan). BxPC-3 and AsPC-1 were maintained in 10% fetal bovine serum (FBS, Biological Industries, Israel)-supplemented RPMI- 1640 medium (Invitrogen, CA, USA). MIA PaCa-2 and PANC-1 were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) with 10% FBS plus 2.5% horse serum (Biological Industries) and 10% FBS, respectively. The four cell lines were cultured at 37oC in a humidified 5% CO2 environment.
Antiserum against thioredoxin-human LC3 was rinsed in rabbits, and anti-human LC3 antibody was purified by affinity chromography on a glutathione-S-transferase-immobilized LC3- Sepharose column. The antibody did not cross-react with GATE- 16 or GABARAP, the other mammalian Arg8 homologues. Affinity purified polyclonal rabbit antibodies against human Atg7 and Atg3 was described. Antiserum against glutathione-S-transferase-human beclin143 was rinsed in rabbits, and the antibodies are purified by affinity chromography on a thioredoxin-beclin 1-immobilized Sepharose column. Monoclonal mouse anti-green fluorescent protein antibody was purchased. Polyclonal rabbit antibodies against mTOR, p70 S6 kinase, phoospho-p70 S6 kinase were also purchased.
Whole cell lysates were extracted from treated cells grown on 6-well plates. Cells were washed in cold PBS and lysised in NP-40, Tris 10 mM pH 8.0, 60 mM KCl, 1 mM EDTA pH 8.0, 1.0 mM DTT, 10μl/ml Protease Inhibitor Cocktail (Sigma P8340)/1ml of lysis buffer and 10mM PMSF. Samples are then placed on ice for 10 mins and the cell lysate collected after centrifugation (13000rpm, 5 mins at 4°C). Total cellular protein was determined by means of the BCA Protein Assay Kit (Pierce). Equal amounts of protein (50μg) are subjected to SDS polyacrylamide gel electrophoresis on 8-12% gels before being trans-blotted onto Immobilin P (Millipore) membranes. Western blotting was performed using antibodies to LC3 (1/1000, Sigma-Aldrich) and beta-Actin (1/5000, Sigma), followed by incubation with the appropriate horseradish peroxidase-conjugated secondary antibodies (Cell Signaling). Signals are detected using ECL™ (Pierce). Autophagy was assessed by western blotting, using an antibody against the autophagy marker LC3 (1/1000, Sigma-Aldrich). As a positive control for the induction of autophagy, cells were starved for 2 hours in Earl’s Whole cell lysates were extracted from treated cells grown on 6-well plates. Cells were washed in cold PBS and lysised in NP-40, Tris 10 mM pH 8.0, 60 mM KCl, 1 mM EDTA pH 8.0, 1.0 mM DTT, 10μl/ml Protease Inhibitor Cocktail (Sigma P8340)/1ml of lysis buffer and 10mM PMSF. Samples are then placed on ice for 10 mins and the cell lysate collected after centrifugation (13000rpm, 5 mins at 4°C). Total cellular protein was determined by means of the BCA Protein Assay Kit (Pierce). Equal amounts of protein (50μg) are subjected to SDS polyacrylamide gel electrophoresis on 8-12% gels before being trans-blotted onto Immobilin P (Millipore) membranes. Western blotting was performed using antibodies to LC3 (1/1000, Sigma-Aldrich) and beta-Actin (1/5000, Sigma), followed by incubation with the appropriate horseradish peroxidase-conjugated secondary antibodies (Cell Signaling). Signals are detected using ECL™ (Pierce). Autophagy was assessed by western blotting, using an antibody against the autophagy marker LC3 (1/1000, Sigma-Aldrich). As a positive control for the induction of autophagy, cells were starved for 2 hours in Earl’s
The assessment of autophagosome number by electron microscopy requires considerable specialized expertise and was becoming increasingly replaced by light microscopic and biochemical methods that are more widely accessible to researchers in different fields. As noted above, the mammalian autophagy protein, LC3, was a marker of autophagosomes. Among the four LC3 isoforms, LC3B is most widely used. Soon after synthesis, nascent LC3 was processed at its C terminus by Atg4 and becomes LC3-I, which had a glycine residue at the C-terminal end. LC3-I was subsequently conjugated with phosphatidylethanolamine (PE) to become LC3-II (LC3-PE) by a ubiquitination-like enzymatic reaction. In contrast to the cytoplasmic localization of LC3-I, LC3-II associates with both the outer and inner membranes of the autophagosome. After fusion with the lysosome, LC3 on the outer membrane was cleaved off by Atg4 and LC3 on the inner membrane was degraded by lysosomal enzymes, resulting in very low LC3 content in the autolysosome. Thus, endogenous LC3 or GFP-LC3 was visualized by fluorescence microscopy either as a diffuse cytoplasmic pool or as punctate structures that primarily represent autophagosomes.
Autophagy (macroautophagy), a well conserved mechanism by which cells adapt to stress such as starvation, had also been recently associated with resistance to cancer therapies. Therefore we investigated whether increased autophagy was involved in the resistance to gemcitabine in the BxPC-3 and Capan-1 cell lines. Total cellular protein is extracted from the different cell lines and sorted CSC following gemcitabine treatment (25 nM, 24hr) and assessed for the expression of LC3 II, a protein associated with autophagosome formation and a marker of autophagy. Autophagy was assessed by western blotting, using an antibody against the autophagy marker LC3 (1/1000, Sigma-Aldrich). As a positive control for the induction of autophagy, cells were starved for 2 hours in Earl’s balanced salt solution (EBSS). Bafilomycin A1 (20 nM, Sigma) was used as an inhibitor of autophagy. Signals are detected using ECL™ (Pierce).
The data were expressed as the mean ± SD and differences between groups were evaluated by Student’s t-test. In all statistical analyses, results were considered to be statistically significant when P-value was less than 0.05.
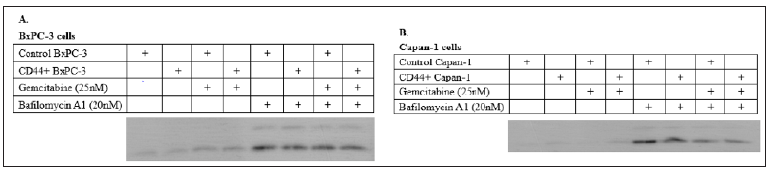
We examined the difference in intensity level of LC3- I or II expression in both CD44+/- BxPC-3 cells and Capan-1 cells of pancreatic cancer cell lines with/without Gemcitabine (25nM) or Bafilomycin A1 (20nM) . (Figure 1) The results showed that LC3-II but not LC3-I strongly expressed in the BxPC-3 and Capan-1 cells when Bafilomycin A1 was added.
Figure 1: Expression levels of LC3‑I, LC3‑II proteins in (A) BxPC-3 and (B) Capan-1 cells treated with 25 nM of gemcitabine with or without addition of bafilomycin A1, 20 nM.
Effect of autophagy inhibitor in combination with gemcitabine on cell viability was performed. BxPC-3 and Capan-1 pancreatic cancer cells were treated with 25 nM gemcitabine or 2- nM bafilomycin A1 alone or in combination for 72 hours respectively was showed in Figure 2. The difference between CD44 CSC and non-CSC had significant difference when only 25 mM was added. It represented that CD44+ pancreatic cancer were more resistant to gemcitabine chemotherapy.
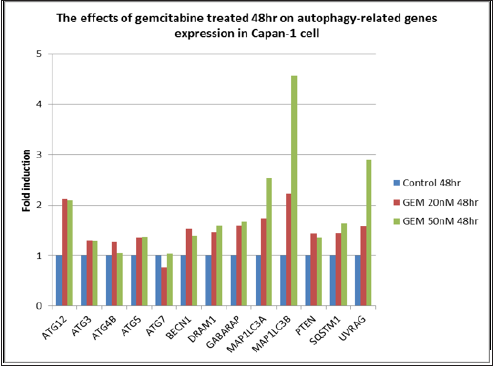
RQ vs Sample with Capan-1 Gemcitabine 50 nM 24 hours were shown in Figures 3 & 4. The ATG12, UVRAG, MAP1LC3B, and MAP1LC3A genes showed significant expressions (1.7 vs 1.0, 1.73 vs 1.0, 2.0 vs 1.0, and 2.1 vs 1.0) . RQ vs Sample with Capan-1 Gemcitabine 50 nM 48 hours were shown in Figures 5 & 6. The ATG12, UVRAG, MAP1LC3B, and MAP1LC3A genes also showed significant expressions (2.2 vs 1.0, 4.6 vs 1.0, 2.55 vs 1.0, and 2.8 vs 1.0). The different expressions with different dosages of Gemcitabine, 20 and 50 nM, were represented in Figures 7 & 8.
Figure 2: Effect of autophagy inhibitor in combination with gemcitabine on cell viability. (A) BxPC-3 and (B) Capan-1 pancreaitc cancer cells were treated with gemcitabine or bafilomycin A1 alone or in combination for 72 hours. The suvival CD+ was significantly higher than CD- with gemcitabine addition, p< 0.05; but the survival of CD+ and CD- had no difference after plus bafilomycin A1.
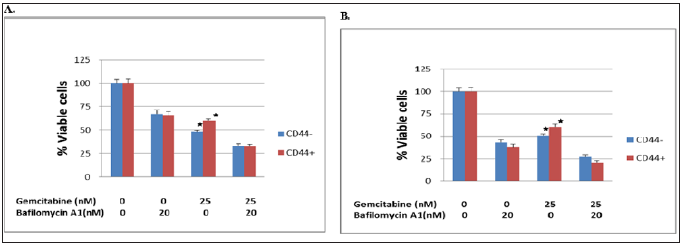
Figure 3: RQ vs Sample with Capan-1 pancreatic cancer cells treated with Gemcitabine 50 nM for 2 hours.
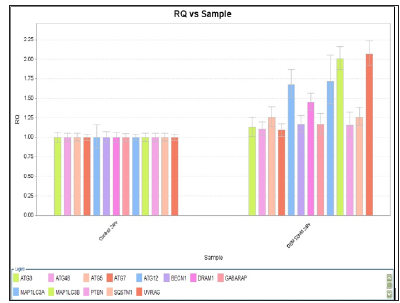
Figure 4: RQ vs Sample with Capan-1 pancreatic cancer cells treated with Gemcitabine 50 nM for 24 hours.
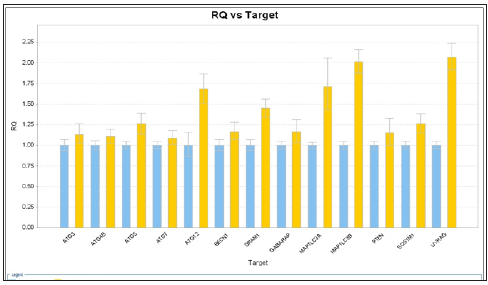
Figure 5: RQ vs Target for autophagy related genes expression with Capan-1 pancreatic cells after gemcitabine 50 nM for 4 hours.
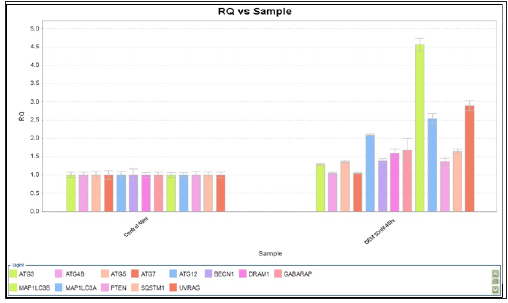
Figure 6:RQ vs Target for autophagy related genes expression with Capan-1 pancreatic cancer cells after gemcitabine 50 nM for 48 hours
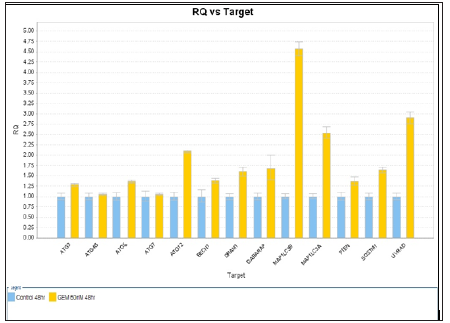
Figure 7: The effects of gemcitabine, 20 and 50 nM, treated for 24hr. on autophagy-related genes expression in Capan-1 pancreatic cancer cells.
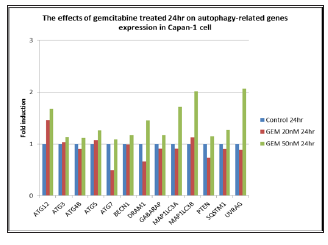
Figure 8: The effects of gemcitabine, 20 and 50 nM, treated for 48hr. on autophagy-related genes expression in Capan-1 pancreatic cancer cells.
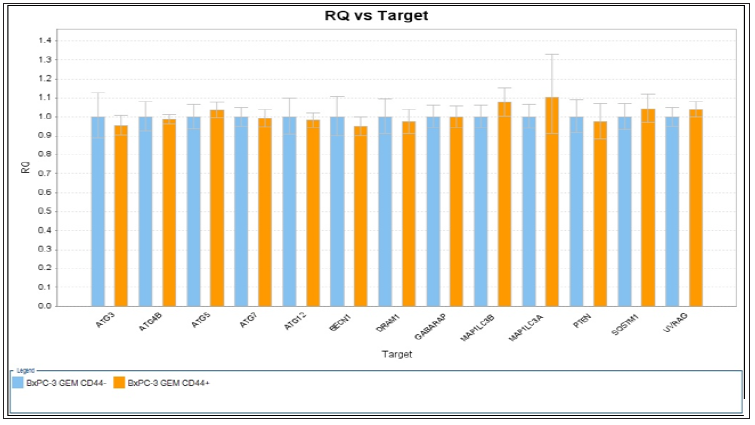
Cells and Non-stem Cells: Effect of autophagy inhibitor in combination with gemcitabine on cell viability was performed. CD44+/- BxPC-3 cells were treated with gemcitabine or bafilomycin A1 alone or in combination for 48 hours respectively was showed in Figures 9 & 10. Mild elevations of MAP1LC3B and MAP1LC3A genes (1.1 vs 1.0 and 1.08 vs 1.0) were found among them. The difference between CSC and non-CSC had no difference.
Figure 9: RQ vs Target for autophagy related genes expression with CD44+/- BxPC-3 pancreatic cancer cells after gemcitabine 20 nM for 48 hours.
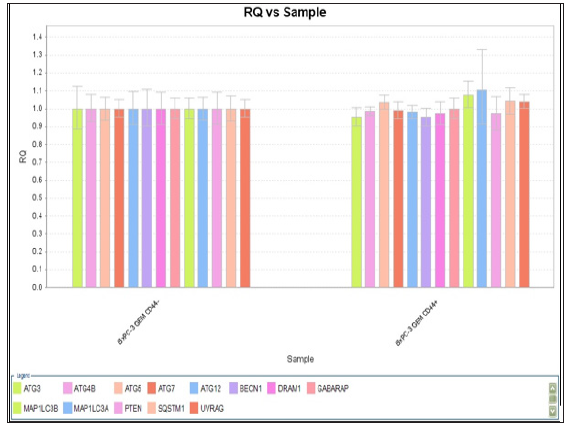
Figure 10: RQ vs. Target for autophagy related genes expression with CD44+/- BxPC-3 pancreatic cancer cells after gemcitabine 20 nM for 48 hours.
As we know that malignancy may result from an increase in cell number due to the disruption of the delicate balance between cell proliferation and elimination. Under certain circumstances, autophagy functions as a safeguard mechanism that promotes uncontrolled cancer cell growth, the up-regulation of autophagic activity is often found in malignant tumors. Autophagy suppresses cancers by buffering metabolic stress when apoptosis is inhibited, to prevent necrotic cell death. Basal autophagy is significantly elevated compared with non-transformed human pancreatic ductal and other tumor cells, confirmed by monitoring LC3 spot formation in PDAC cell lines and primary tumors [29]. Knockdown of autophagy genes such as ATG5 and ATG3 by RNA interference significantly inhibits cell growth and colony formation of PDAC cell lines in vitro. [30-31]. Our data in Figure 1 showed the expression of LC3-II significantly increased when bafilomycin was added which prevented the blockage of LC3-I both in BxPC-3 cells and Capan-1 cells. The difference between CD44+ and control non-stem cells had no difference. The autophagy activity between stem and non-stem cells could not showed significant difference. However, the percentage of viable cells for CD44+ stem cells was higher and weaker autophagy after gemcitabine was added in both BxPC-3 and Capan-1 cells (Figures 2 & 3).
The difference could demonstrate that resistance of gemcitabine treatment in CD44+ stem cells may be due to its higher autophagy activity. The suppression of autophagy through cancer stem cells of PDAC may prevent the drug resistance of gemcitabine during chemotherapy for PDAC. Autophagy is strictly regulated by a limited number of autophagy-related genes (ATGs) which were originally discovered in yeast, and over 20 of them have been identified in mammalian cells [33]. Some key autophagic mediators, such as ATGs, P13K, mTOR, p53, and Beclin-1 might be implicated in modulating autophagic activity in cancer initiation and development. The autophagic mediators and their corresponding proteins can integrate into cancer cell signaling networks and ultimately regulate cell survival or death [34]. ATGs play significant roles in autophagosome formation and autophagy regulation. They have numerous links with many human diseases, particularly cancers (35). Autophagy regulates several ATGs in a two-faced manner, such as Atg1/ULK1 (Unc51-like kinase), Atg4, Beclin-1/ Atg6 and LC3/Atg8, which may determine the fate of cancer cells, at least to some extent.
Some regulators are able to synchronously influence apoptosis and autophagy, despite the remarkable differences between these two types of programmed cell death. Autophagy can protect tumors from cancer therapeutics and prevent apoptotic cell death via the elimination of damaged cellular contents. Apoptosis is mainly responsible for the physiological elimination of cells, which is regulated by a variety of cellular signaling pathways and closely associated with cancers (35). Strong expression of LC3 is associated with enhanced expression of the hypoxia marker carbonic anhydrase IX at the peripheral area, which correlates with poor outcome and a short disease-free period. Another study showed that LC3 is low or absent in normal exocrine pancreas and in low-grade PanIN-1 and PanIN-2 lesions, whereas staining is upregulated and exhibits a vesicular staining pattern in all high-grade PanIN-3 and PDAC. Our results also revealed significant expression of ATG12, UVRAG, MAP1LC3B, and MAP1LC3A genes in pancreatic cancer cell lines after gemcitabine treatment.
The four genes might provide clues to relate to the drug resistance through autophagy pathway. The difference of MAP1LC3B, and MAP1LC3A genes was also demonstrated between CD44+ and CD44-, which indicated that drug resistance of cancer stem cells, such as CD44+ may be inhibited through autophagy blockage. Considering the complex relationship between autophagy and cancer, any clinically oriented application of autophagy inducers or inhibitors must be conducted with extreme caution because both of the agents might backfire and promote tumorigenesis. Inhibition of protective autophagy might break the resistance to the detrimental micro-environment and lead to cell death. However, undesirable results, such as necrosis and inflammation, are also possible, which make autophagy inhibition more detrimental than beneficial. Due to the complex relation-ship between autophagy and cancer and the potential danger in the application of autophagy-targeting drugs in cancer therapeutics, it is important for molecular therapy to identify drugs targeting the mechanisms of autophagy based upon the network, combining experimental approaches and computational methods under the guidance of systems biology.
Several such challenges facing this field are how to cause tumor cells to die after triggering autophagy using several known antitumor drugs, how to eliminate autophagic protective functions toward tumor cells, and how cancer cells can balance between several conflicting signals. To solve these problems, systems biology may be the key point in accomplishing these goals in the everlasting battle against cancer. In conclusions, the percentage of viable cells for CD44+ stem cells was higher and weaker autophagy after gemcitabine was added in both BxPC-3 and Capan-1 cells. The difference of MAP1LC3B, and MAP1LC3A genes was also demonstrated between CD44+ and CD44-. Blockage of such genes might contribute to the effectiveness of Gemcitabine chemotherapy in the future. We hope our further study can reveal the actual role of autophagy in drug resistance of pancreatic cancer and hope to improve the chemotherapeutic effects and survival of pancreatic cancer patients.
This work was supported by a grant (NSC-101-2314-B-182- 027-MY2) from the National Science Council, and grant (CMRPG3D0081) from Chang Gung Memorial Hospital, Taiwan.


