Abstract
This study investigated the prevalance of New Delhi Metallo-β-Lactamase (NDM) resistant gene in 57 E. coli collected clinical isolates from hospitals in Khartoum state, Sudan. An emerging carbapenemase, New Delhi metallo-lactamase (NDM), was first reported in a Swedish patient with a hospitalization history in India, it exhibited resistant to all β-lactams except for monobactams, and has great potential to cause global health crisis [1]. Initially, blaNDM gene was endemic to India subcontinent and NDM-producing isolates tested worldwide have geographical links with these high prevalence areas. Out of 200 Escherichia coli collected from clinical samples; 57 resistant strains were identified in different Khartoum state hospitals (n= 57). These resistance isolates were processed by bacteriological conventional methods and then were tested by antimicrobial profile and concentrated on Meropenem and Imipenem antibiotics and then RT-PCR with β-lactamase NDM primer were done. The NDM positive E.coli were 48 (80.7%) from total (n=57) resistant isolates. Within the positive blaNDM; 40.3% were resistant to Imipenem, 59.6% sensitive to Imipenem. blaNDM positive showed sensitivity profile to Meropenem; 38.5% of them were resistant to Meropenem, 61.5% were sensitive to Meropenem. The MHT showed that 56.2% of the blaNDM positive organisms were positive for Imipenem MHT and 64.9% were positive for Meropenem MHT. Our findings provides additional evidence that the majority of the circulating clinical isolates Escherichia coli in Khartoum hospitals in Sudan harbouring NDM antibiotic resistance gene.
Keywords: Resistant; Imipenem; Meropenem; NDM β-lactamase; Escherichia Coli
Introduction
Carbapenem, a β-lactam that is highly potent against Gramnegative bacteria, has been recognized as a last resort for treating of infections caused by multidrug-resistant bacteria. However, the increasing number of Carbapenem-Resistant Enterobacteriaceae (CRE) is unexpected despite infection control efforts, and it poses a great challenge to clinic [2]. Carbapenem resistance is predominantly attributed to the presence of carbapenemases, among which Class A (blaKPC), Class B (blaNDM, blaVIM, blaIMP), and Class D (blaOXA-48) types are most common for Enterobacteriaceae [3,4]. New Delhi Metallo-beta-lactamase (NDM) is a plasmid encoded transferable molecular class B β-lactamase gene. Unlike class A, C and D beta-lactamases, NDM has Zinc ions at its active site, and it can hydrolyze all beta-lactam antimicrobials except for monobactam [5]. Moreover, most NDM positive bacteria are resistant to a wide variety of other antimicrobial classes and carry several additional resistance mechanisms for example to aminoglycosides, fluoroquinolones, macrolides and sulfonamides, leaving few treatment options available [6].
NDM was first discovered in 2008 by Swedish scientists treating a patient of Indian descent who travelled to India, where he was hospitalized, initially in the Punjab area, but then in New Delhi, for the management of a gluteal abscess. In January 2008 he was admitted to a hospital in Orebro, Sweden, where, on the day after admission, a urine culture yielded an isolate of Klebsiella pneumoniae that was resistant to multiple antibiotics including Carbapenems (ertapenem, Imipenem and Meropenem) [7]. This strain was not isolated from any subsequent cultures, but stool samples tested following transfer of the patient to a nursing home in March 2008 yielded a Carbapenems resistant strain of Escherichia coli. Phenotypic testing of both isolates suggested that the Carbapenems resistance was due to the production of a Metallo- β-Lactamase (MBL), but PCR analysis failed to detect known MBL genes. The global dissemination of carbapenem resistance has recently received much attention especially following reports of the international spread of Enterobacteriaceae strains producing carbapenem mases such as KPC and NDM from endemic areas [8]. This dissemination is facilitated by increased international travel, which has been described as a risk factor for the acquisition of infections due to drug resistant organisms such as CTX-M-producing E. coli, as well as various carbapenemase-producing Gram-negative bacteria, including NDM and KPC producers [9].
Material and Methods
This study was conducted at the Khartoum teaching Hospital, Bahri, and Omdurman hospitals. Receiving referrals from 3 districts and serving a population of more than sex million) in the period between December 2016 to November 2018.
Bacterial Isolates
A total of 200 Escherichia coli isolates were collected from 700 different clinical specimens (185 urine, 105 sputum, 235 wound, 100 bloodstream infections, and 75 stool samples) as part of the routine hospital laboratory procedure. Only 57 Isolates were identified as carbapenem-resistant clinical isolates by using standard laboratory methods including bacteriological and biochemical tests such as Gram stain, growth on EMB medium, indole test and motility test and also KIA test. The identified Eschrichia coli isolates were stored at-70°C in trypticase soy broth (Merck, Darmstadt, Germany) supplemented with 10% glycerol and 2g of charcoal until further investigation needs. Escherichia coli ATCC 25922 was used as the reference strain throughout the study. A set of 143 carbapenemsensitive clinical isolates were used as negative controls.
Antimicrobial Susceptibility Testing
The enzymatic tests of meropenem and imipenem were done after diffusion method that measured on Muller Hinton Agar plates (Oxoid, Basingstoke, UK) by the agar diffusion method and interpreted according to CLSI [10]. Meropenem and Imipenem discs were purchased from Himedia, Mumbai (India). Antimicrobial profile against Escherichia coli drugs was done for the carbapenemresistant isolates using disc-diffusion method according to CLSI [10].
DNA Extraction and Sample Preparation
DNA extraction was performed using iNtRON genomic DNA extraction kit. The isolates were cultivated in nutrient agar and incubated at 37OC overnight aerobically. A 300 μl of heavy organism suspension was prepared in a 1.5ml tube to be extracted. The kit consists of washing buffer, lysis buffer, protein precipitation solution and rehydration solution as well as 70% Ethanol. All the materials were brought to room temperature before use.
Extraction Protocol
A volume 900μl of washing solution was added to the prepared sample to remove all media artifacts and metabolic products of the organism. Vortexed sufficiently the mixture was then incubated for 10min on ice and centrifuged at 13,000rpm for 30sec to obtain clean cell pellet. The supernatant solution was removed, and the cell pallet was suspended and 600μl of Lysis Buffer was added to the tube. To enhance the lysis, the lysate was incubated in a sonication water bath at 65c˚ and frequency of 50KHZ for 10 minutes. To prevent the protein contamination, 200μl of Protein PPT Buffer was added and vortexed for 10 secs, then incubated in ice bath for 5min. After incubation, the samples were centrifuged at 13,000rpm for 5 min at room temperature. The supernatant was then transferred to a fresh 1.5 tube and 600μl of isopropanol was added and mixed well then, the mixture was centrifuged at 13,000rpm for 1minutes at room temperature. To precipitate the DNA, 600μl of 70% Ethanol was added and gently mixed, the tubes were then centrifuged at 13,000rpm for 1 min at room temperature. Then the Ethanol was removed, and the DNA pallet was dried. For DNA rehydration 100μl of DNA Rehydration Solution was added to the tubes. All samples were stored at -20c.
Reaction Setup
The test reaction was set to a total volume of 25μl. 5μl master mix, 1μl of forward and reverse primers, the amount of Eva green dye used in the reaction varied per the amplicon size; 1μl Eva green was used for blaNDM as it was the smallest amplicon size. 0.3 μl of the DNA template was used and the rest of the reaction’s volume was double distilled water. Pooling volume per the tested samples number was done to minimize pipetting errors. The test was conducted using 0.2ml PCR tubes.
Real-Time PCR Conditions
Using Sacycler 96 instrument of Sacace biotechnology, real-time PCR protocol was carried out under the following: Hot start 94Cᵒ for 10 min; with number of cycles 40 and the loop steps were as follow: Denaturation at 94Cᵒ for 45 seconds, annealing at 52Cᵒ for 45 seconds and elongation 72Cᵒ for 30 seconds which included the fluorescent data at acquisition 533nm filter, a final elongation step was set to be at 72C˚ for 10 min. After the end of the polymerization cycles; melting curve step was added to start from 65Cᵒ gradually increasing by 0.1C/s to 95C, with the fluorescence data acquisition every 1 second.
Results
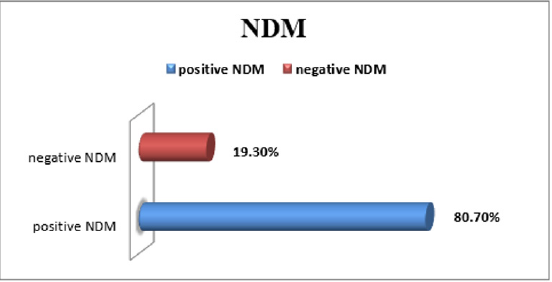
In the blaNDM; 80.7%of the samples were positive (Figures 1 & 2). Strong positive samples were indicated by the concentration of the PCR product and fluorescent reading.
Amplification protocol:
Volume (35mcL)
1. 94.0°C - :10:00
2. 94.0°C - :00:30
52.0°C - :00:45 *36
72.0°C - :00:50
3. 70.0°C - :00:15 *230 (Every 0.10 sec)
blaNDM and Antibiotics
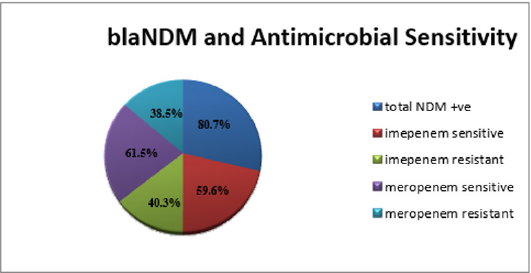
Within the positive blaNDM; 40.3% were resistant to Imipenem, 59.6% sensitive to Imipenem (Figure 3).
blaNDM positive showed sensitivity profile to Meropenem; 38.5% of them were resistant to Meropenem, 61.5% were sensitive to Meropenem (Figure 3).
blaNDM and Enzyme Assay
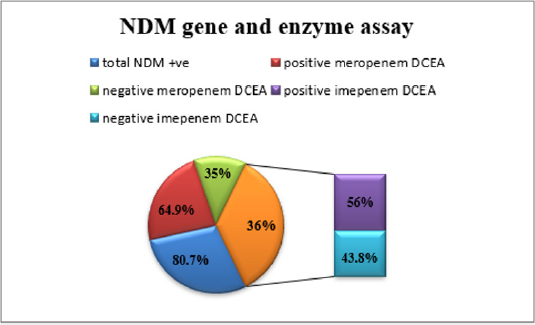
Enzymes Assays reveled the following; 64.9% of the blaNDM positive organisms were positive in the Meropenem enzyme assay, and 56% were positive for the Imipenem enzyme assay (Figure 4).
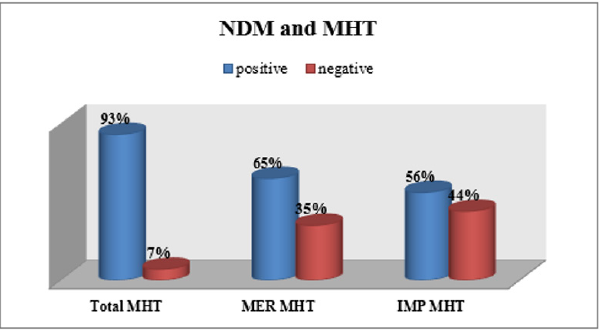
blaNDM and MHT
The MHT showed that 56.2% of the blaNDM positive organisms were positive for Imipenem MHT and 64.9% were positive for Meropenem MHT (Figure 5). 80.7% of the samples were positive for blaNDM, 56% of them were resistant for Imipenem and 44% sensitive. 35% of them were sensitive to Meropenem and 65% were resistant to it. 61% of these organisms were positive for the enzyme assay if the three substrates. 44% of the organisms were negative for Imipenem MHT and 65% were positive for Meropenem MHT.
Discussion
Our findings emphasized the epidemiological characteristics and the high resistance of carbapenemase-resistant E. coli. The blaNDM is the prevalent mechanism of carbapenem-resistant E. coli, and could be transferred by plasmid-mediated horizontal gene transfer in Khartoum, Sudan. The surveillance of antimicrobial resistance was remain as a major objective for the increased in the prevalent of multidrug resistance and a greatest concern for carbapenem-resistant E. coli associated with different infections. In general data on the dissemination of antimicrobial genes on reports of emerging (IPM, VIM and NDM) MBL genes among Gram-negative strains have been published worldwide, including sub-Saharan Africa and Middle East countries such as Ethiopia [12], Kenya [13], Uganda [14], Egypt [15]. , South Africa [16], Saudi Arabia [17], Iraqi [18], Kuwait [19], and Lebanon [20], were agreed with our study and were found a high rate of bla-NDM gene. In the same our view many studies were found increased rate of NDM as Kumarasamy and colleagues [21], provide compelling evidence that NDMproducing Enterobacteriaceae (mostly K. pneumoniae and E. coli) are widespread in India and Pakistan. They also found that many UK patients infected with NDM-producing bacteria had recently traveled to India to undergo several types of medical procedures. The patients presented with a variety of hospital- and communityassociated infections with urinary tract infections (UTIs) being the most common clinical syndrome. Recent reports from the subcontinent (including India, Pakistan and Bangladesh) show that the distribution of NDM β-lactamases among Enterobacteriaceae are widespread through these countries [22-24].
Since 2011, NDM-1-positive bacteria have been reported worldwide [25]. Most are Enterobacteriaceae including E. coli from patients hospitalized in 2009 and 2010 with an epidemiological link to the Indian subcontinent. Recent findings suggest that the Balkan states and the Middle East might act as secondary reservoirs for the spread of NDM-1, which may or may not initially have reached these countries from the Indian subcontinent [25]. Enterobacteriaceae with NDM-1 have been recovered from many clinical settings, reflecting the disease spectra of these opportunistic bacteria, including hospital and community-onset urinary tract infections, septicemia, pulmonary infections, peritonitis, device-associated infections and soft tissue infections. NDM-1-positive bacteria have been recovered from the gut flora of travelers returning from the Indian subcontinent and undergoing microbiological investigation for unrelated diarrheal symptoms [26]. There is also widespread environmental contamination by NDM-1-positive bacteria in New Delhi [27]. Cuong Q et al were found out of 100 strains from clinical samples (50 isolates of E. coli), 47 isolates were positive by real-time PCR. In those positive strains, 36 carried blaNDM. 58.3% (21/36) strains carrying blaNDM. finally that research indicates the lower prevalence of resistant strains of Escherichia coli in clinical isolates from different hospitals in Southern Vietnam [28].
Conclusion
The outcome of this work revealed that in Sudan must be take care and doing alarming mark for hazard future of these bacteria in soon time in mind the emergence of these Escherichia coli resistant strains. Also molecular detection by PCR techniques for detection of all genes responsible for resistant E.coli infections is highly recommended to have clear idea about genetic variation in E.coli resistant strains. All of these studies [25-28] were agreed with our research in found high rate of NDM but were less our results obtained; gerographical distribution and sample size were different. While NDM gene was present with high frequency (80.7%), putting in mind consideration the difficulties facing the detection of specific resistant genes in traditional most common clinical microbiology laboratory, it recommended to follow the CDC policy which adopted criteria to identify Escherichia coli resistant strains. Further organized surveillance is needed for perfect knowledge about specific determination for spreading of resistant genes in Sudan. And an effort should be made with governmental authorities to control abuse and randomization used of antibiotics and prevention of emergence to resistant strains of Escherichia coli.
References
- Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, et al. (2009) Characterization of a new metallo-beta-lactamase gene, bla (NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother 53(12): 5046-5054.
- Zilberberg MD, Shorr AF (2013) Secular trends in gram-negative resistance among urinary tract infection hospitalizations in the United States, 2000-2009. Infect Control Hosp Epidemiol 34(9): 940-946.
- Walsh TR (2010) Emerging carbapenemases: A global perspective. Int J Antimicrob Agents 36(Suppl. 3): S8-S14.
- Albiger B, Glasner C, Struelens MJ, Grundmann H, Monnet DL (2015) Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries. Euro Surveill 20(45): 30062.
- Saini A, Bansal R (2012) Insights on the structural characteristics of NDM-1: The journey so far. 2(04): 323-334.
- Kim Y, Tesar C, Mire J, Jedrzejczak R, Binkowski, et al. (2012), Structure of apo-and monometalated forms of NDM-1-a highly potent carbapenem-hydrolyzing metallo-β-lactamase. PloS one 6(9): e24621.
- Krizova L, Bonnin RA, Nordmann P, Nemec A, Poirel L (2012) Characterization of a multidrug-resistant Acinetobacter baumannii strain carrying the blaNDM-1 and blaOXA-23 carbapenemase genes from the Czech Republic. Journal of Antimicrobial Chemotherapy 67(6): 1550-1552.
- Nordmann P, Naas T, Poirel L (2011) Globalspread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17(10): 1791-1798.
- Van der Bij AK, Pitout JD (2012) The role of international travel in the worldwide spread of multiresistant Enterobacteriaceae. J Antimicrob Chemother 67(9): 2090-2100.
- Wayne P (2002) National committee for clinical laboratory standards. Performance standards for antimicrobial disc susceptibility testing.
- Monteiro J, Widen RH, Pignatari AC, Kubasek C, Silbert S (2012) Rapid detection of carbapenemase genes by multiplex real-time PCR. Journal of Antimicrobial Chemotherapy 67(4): 906-909.
- Pritsch M, Zeynudin A, Messerer M, Baumer S, Liegl G, et al. (2017) First report on blaNDM-1-producing Acinetobacter baummanii in three clinical isolates from Ethiopia. BMC Infect Dis 17: 180.
- Poirel L, Revathi G, Bernabeu S, Nordmann P (2012) Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob Agents Chemother 55(2): 934-936.
- Zafer MM, Al-Agamy NH, El-Mahallawy HA, Amin MA, Ashour MS (2014) Antimicrobial resistance pattern and their β-lactamase encoding genes among Pseudomonas aeruginosa strains isolated from cancer patients. Bio Med Research Int 2014: e101635.
- Diab M, Fan N, El-Said M, El-Dabaa E, El-Defrawy I, et al. (2013) Occurrence of VIM-2 Metallo-β-lactamases in imipenem resistant and susceptible Pseudomonas aeruginosa clinical isolates from Egypt. Afri J Microbiol 7(35): 4465-4472.
- Sigh-Moodley A, Perovic O (2016) Antimicrobial susceptibility testing in predicting the presence of carbapenemase genes in Enterobacteriaceae in South Africa. BMC Infect Dis 16(1): 536.
- Shaik S, Fatima J, Shakill S, Rizvi SMD (2015) Antibiotic resistance and extended-spectrum beta-lactamases: types, epidemiology, and treatment. Saudi Biol Sci 22(1): 90-101.
- Anoar KA, Ali FA, Omer SA (2014) Detection of Metallo β-lactamase enzyme in some gram-negative bacteria isolated from burn patients in Sulaimani city, Iraq. European Scientific J 10(1): 485-496.
- Jamal W, Rotimi VO, Albert MJ, Khodakhast F, Nordmann P, et al. (2013) High prevalence of VIM-4 and NDM-1 Metallo-beta-lactamase among carbapenem-resistant Enterobacteriaceae. J Med Microbiol 62(8): 1239-1244.
- El-Herte RI, Araj GF, Mata GM, Baroud M, Kanafani ZA, et al. (2012) Detection of carbapenem-resistant Escherichia coli and Klebsiella pneumonia producing NDM-1 in Lebanon. J Infect Dev Ctries 6(5): 457-461.
- Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, et al. (2010), Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10(9): 597-602.
- Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, et al. (2011) Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006-2007. Antimicrob Agents Chemother 55(3): 1274-1278.
- Castanheira M, Mendes RE, Woosley LN, Jones RN (2011) Trends in carbapenemase-producing Escherichia coli and Klebsiella spp. from Europe and the Americas: report from the SENTRY antimicrobial surveillance programme (2007-09). J Antimicrob Chemother 66(6): 1409-1411.
- Lascols C, Hackel M, Marshall SH, Hujer AM, Bouchillon S, et al. (2011) Increasing prevalence and dissemination of NDM-1 metallo-beta-lactamase in India: data from the SMART study (2009). J Antimicrobial Chemother 66(9): 1992-1997.
- Johnson AP, Woodford N (2013) Global spread of antibiotic resistance: The example of New Delhi metallo-beta-lactamase (NDM)-mediated carbapenem resistance. Journal of medical microbiology 62: 499-513.
- Leverstein-Van Hall MA, Stuart JC, Voets GM, Versteeg D, Tersmette T, et al. (2010) Global spread of New Delhi metallo-beta-lactamase 1. Lancet Infect Dis 10(12): 830-831.
- Walsh TR, Weeks J, Livermore DM, Toleman MA (2011) Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11(5): 355-362.
- Cuong QH, Hai DN, Huy QV, Anh TN, Bin T, et al. (2019) Emergence of New Delhi Metallo-Beta-Lactamase (NDM) and Klebsiella pneumoniae Carbapenemase (KPC) Production by Escherichia coli and Klebsiella pneumoniae in Southern Vietnam and Appropriate Methods of Detection: A Cross-Sectional, Study. 9757625. 9.

 Research Article
Research Article