Abstract
The association between Fibroblast Growth Factor Receptor 2 (FGFR2) gene polymorphism and expression with breast cancer varies with ethnicity and geographical distribution. Several studies have been conducted with different genotypes of this particular gene polymorphism among various ethnicities throughout the world. The main purpose of this study was to evaluate the relationship between FGFR2 (rs2981582) gene polymorphism and FGFR2 gene expression with breast cancer in Indian women and also to establish the relative gene expression with different genotypes of the polymorphism, if there is any. Method: A total of 75 cases were selected, out of which 25 were malignant cases,25 healthy controls and 25 benign cases. Restriction Fragment Length Polymorphism Polymerase Chain Reaction (RFLP PCR) was used to determine FGFR2 Single Nucleotide Polymorphism and Real Time Quantitative Polymerase Chain Reaction (RT qPCR) for FGFR2 gene expression. The strength of association was estimated using Odds Ratios (ORs) and 95% Confidence Intervals (CIs). RESULTS: Our results showed that FGFR2 (rs2981582) polymorphism is not significantly associated with breast cancer in Indian women. However, FGFR2 gene expression is significantly increased in breast cancer but the differential gene expression associated with different genotypes (TT / TC / CC) is not significant in both malignant and benign cases. Therefore, we conclude from this study that FGFR2 rs2981582 SNP does not influence the expression of FGFR2 in this population.
Introduction
Breast Cancer (BC) is one of the most common malignant tumors among women all over the world and has the highest mortality rate amongst the cancers afflicting women [1]. In recent years, its incidence has increased among young women with increasing tendency of chemo-resistance and recurrence [2,3]. Genetic factors such as BRCA1, BRCA2, HER2, CyclinD1, MAP3K1, FGFR2, TOX3, etc play a major role in etiopathogenesis of breast cancer [4]. Lots of researchers have reported the association between FGFR2 polymorphism and BC risk [5-7]. FGFRs mediate signaling of fibroblast growth factors (FGFs) and have numerous important functions [8]. FGFR2 gene is located at chromosome 10q26 and acts through multiple downstream signaling pathways that play vital roles in cell proliferation, survival and differentiation [9]. The role of FGFR pathway as a predictive/prognostic marker has been investigated through various studies. According to these studies, aberrant FGFR2 expression is associated with an increased risk of BC and correlates with poor prognosis [10,11]. According to Gene Wide Association Study (GWAS), five single nucleotide polymorphisms (SNPS) (rs7895676, rs2912781, rs10736303, rs2912778 and rs2981582) in the non-coding region of FGFR2 were found to have a significant association with breast cancer. FGFR2 (rs2981582) in intron 2 has been identified as the most significant breast cancer risk locus by the Breast Cancer Association Consortium genome wide association study [12]. Hunter et.al (2007) also identified four other SNPs, rs11200014, rs2420946, rs1219648 and rs2981579 in intron 2 of FGFR2 that are also associated with breast cancer [13]. Therefore, it becomes very important to understand the FGFR pathway on how it leads to BC and how it can be targeted. Several trials are being done with multiple agents that target FGFR pathway components [14-16]. According to these trials, identification of patients with FGFR pathway amplified tumors who may respond to this treatment may be possible. Large number of studies are available on relevance between FGFR2 single nucleotide gene polymorphisms (rs2981582 and rs2981579) and breast cancer, but the results vary with geographical differences [17]. There is hardly any study exploring the association of rs2981582 FGFR2 polymorphism in patients of breast cancer with Indian ethnicity. Therefore, we developed an interest to elucidate the frequency of this polymorphism and its influence on FGFR2 expression in Indian patients with breast cancer.
Materials and Methods
Study Site and Patient Recruitment
The present study was conducted at the Department of Biochemistry in collaboration with the Department of Surgery and Department of Pathology, Maulana Azad Medical College and Associated Hospitals, New Delhi, India. The Institutional Ethics Committee (IEC) of Maulana Azad Medical College approved the study and written informed consent was provided by all participants for blood sampling and genetic testing. All the research has been conducted in accordance with the Helsinki declaration. Patients were consecutively recruited from July, 2018 to December, 2020 in Lok Nayak Hospital, New Delhi, India. Twenty-five female patients who were histopathologically diagnosed with breast cancer were included in the study. Another 25 patients with benign fibroadenoma were also recruited. The control subjects comprised of twenty-five unrelated healthy women. Patients with history of other malignancies or other systemic diseases were excluded from the study.
Genotyping
Peripheral blood samples from cases and healthy control subjects were collected in EDTA anticoagulant vials and stored at −80°C until used for further process. Whole genomic DNA was isolated from the peripheral blood mononuclear cells (PBMNCs) using HiPurATM Blood Genomic DNA Miniprep Purification Kit (Ref: MB504-50PR, Country of origin: India) according to the manufacturer’s instructions. The quality of DNA was checked by Ethidium Bromide (EtBr) stained 1% agarose gel electrophoresis. The DNA quantity and integrity were determined by A260/280 ratio using NanoDrop spectrophotometer (Washington, DC, USA). Polymerase Chain Reaction Restriction Fragment Length Polymorphism (PCR-RFLP) technique was used to study rs2981582 single nucleotide polymorphism in FGFR2 gene. The detailed information of the FGFR2 SNPs, primer sequences used for amplification, PCR amplicon sizes, restriction edonucleases used and size of products obtained after restriction digestion are given in Table 1. The restriction digestion fragments were resolved and analyzed by 3% to 3.5% agarose gel electrophoresis using EtBr as staining dye.
RNA Isolation and cDNA Synthesis
Total RNA was extracted from all the subjects using Geneaid
Total RNA Mini Kit (Cat. No. RB100, Country of origin: Taiwan)
according to the manufacturer’s instructions. Briefly, 300μL of
peripheral blood in an eppendorf was mixed with 900μL of RBC
lysis buffer, incubated at room temperature for half an hour and
centrifuged for 10minutes at 3500 RPM. The supernatant was
discarded, and this step was repeated till a whitish cell pallet
appeared at the bottom of the eppendorf tube. This cell pallet was
mixed with 200μL cell lysis buffer and incubated for 10minutes.
The ingredients of the eppendorf tube were then transferred
to columns, provided with the kit, and centrifuged at high speed
(14000 RPM) for 2minutes. The temperature of the centrifuge
machine was set at 4°C. The columns were then washed with 75%
alcohol and RNA was eluted down in RNAse free water (100μL).
RNA integrity was determined by agarose gel electrophoresis (2%)
and concentration was determined by measuring absorbance at
260 nm as discussed above.
The RNA was converted to complementary DNA (cDNA) using
cDNA synthesis kit (Thermo Fisher Scientific, USA) by following
manufacturer’s instructions as provided with minor modifications.
I brief, total RNA (100 ng), 2 μL oligo (dT) primer, and 9 μL water
were mixed and incubated at 70°C for 5 minutes in a thermal
cycler. This reaction mixture was then mixed with 5X RT buffer
(4 μL), 5 mM dNTPs (2 μL), 1 μL each of RNAse inhibitor, and reverse transcriptase enzyme in a total volume of 20 μL. This mix
was incubated at 42°C for 1 h and then heated to 95°C to stop the
reaction. The cDNA was stored at −80°C until used for expression
studies.
Quantitative Real Time Polymerase Chain Reaction (qRT-PCR)
Quantitative RT-PCR was used to carry out relative expression
of FGFR2 gene on a Rotor-Gene Instrument (QIAGEN; Skelton
House, Lloyd, Manchester, UK). β-actin gene was used as reference
gene for normalizing. The primers used were as follows:
FGFR2 forward primer 5′-TCCACATGGAGATATGGAACAGGA-3′
FGFR2 Reverse primer 5′-GGAGCTATTTATCCCCGAGTG-3′
β-actin Forward primer 5′-CGACAACGGCTCCGGCATGTGC-3′
β-actin Reverse primer 5′-CGTCACCGGAGTCCATCACGATGC-3′
A 20 μL reaction mixture consisting of 10 μL SYBR Green
Master Mix, 1 μL cDNA (equivalent to 100 ng RNA), 0.3 μL forward
primer (25 pm/μL), 0.3 μL reverse primer (25 pm/μL), and 8.4 μL
water. The thermal cycling conditions were standardized to match
both FGFR2 and β-actin specific primers. The final standardized
temperature profile consisted of a denaturation program of 95°C for
5 min followed by 40 cycles of denaturation at 95ºC for 15 seconds,
annealing at 60ºC for 45 seconds, and extension at 72ºC for 50 seconds
with a final extension at 72ºC for 10 minutes. The quantification
was expressed as 2−ΔΔCT or fold change using Livak method adopted
from (https://doi.org/10.1177/1010428317713857 and https://
doi.org/10.1007/s13577-020-00370-6).
Statistical Analysis
Chi-square test was performed for comparison of genotypic
frequency distribution between:
I) All patients and healthy controls,
II) Benign patient’s vs healthy controls and
III) Malignant patient’s vs healthy controls and IV) benign
vs malignant patients. Odds ratio along with corresponding 95%
confidence intervals (CIs) were used for estimation of individual
genotypic risk for breast cancer by employing codominant model
of inheritance. Continuous data was analysed by student t-test and
One-way ANOVA (Analysis of variance). Moreover, all the p-values
were corrected, in case of multiple comparisons, by employing
Tukey’s post hoc test. These statistical analyses were performed by
GraphPad Prism 5.lnk software package and a p value of <0.05 was
considered statistically significant.
Results
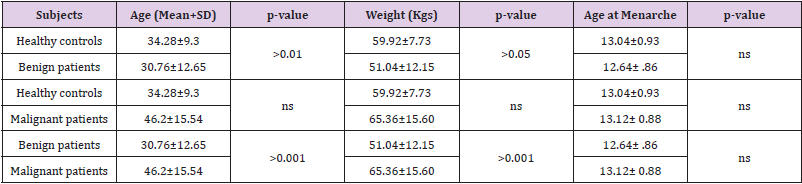
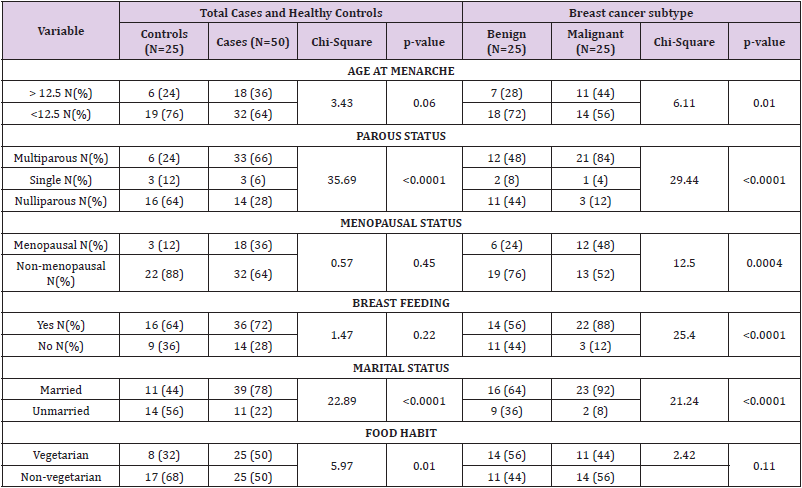
The clinicopathological and demographic characteristics of patients and healthy controls are represented in Tables 1 & 2. In contrast to healthy controls (mean ±SD=34.28±9.3), the breast cancer patients tended to be younger (mean ± SD=32.52+11.13) taken together was more. Further, when the breast cancer patients were subdivided, benign patients were comparatively found to be younger (mean ±SD=30.76±12.65) followed by healthy subjects (mean ±SD=34.28±9.3) and then by malignant patients (mean ±SD=46.2±15.54). Principally, we did not find any significant differences in basic demographic characteristics among the healthy subjects and breast cancer patients when taken together except that breast cancer patients significantly tend to be multiparous and married. However, significant differences were seen in basic characteristics including age at menarchy, parous status, menopausal status, breast feeding and marital status, but feeding habits, between subtypes. All the basic characteristics of healthy and breast cancer subjects, along with statistical analysis, have been presented in Tables 2 & 3.
Risk Analysis of FGFR2 Polymorphisms in Concert with Breast Cancer
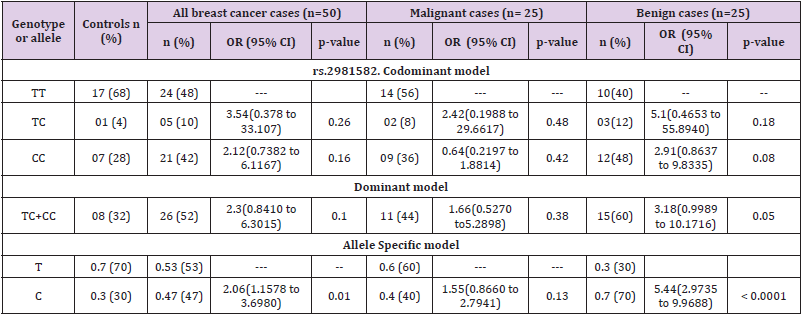
The genotypic and allele distribution of FGFR2 polymorphisms (rs nos. of FGFR2 pols) are shown in Table 3. The risk analysis based on calculation of odds ratio showed association of FGFR2 polymorphism with overall breast cancer risk. It was observed that FGFR2 polymorphism exhibited a heightened overall breast risk in both codominant and dominant models though it was not significant. In codominant model the heterozygous genotype (TC) depicted the highest risk (OR=3.54; 95% CI= 0.378 to 33.107; p=0.26) followed by recessive homozygous (CC) (OR=2.12; 95% CI= 0.7382 to 6.1167; p=0.16); dominant model; TC+CC; (OR=2.30; 95% CI= 0.8410 to 6.3015; p=0.10). The allele specific model depicted a statistically increased risk of overall breast cancer associated with the minor allele of the polymorphism (C allele; OR=2.06 95% CI=1.1578 to 3.6980; p=0.01).When patients were sub-grouped based on the type of tumor; benign and malignant, TC genotype in dominant model, TC+CC additive genotype in dominant model and C allele in allele specific model showed a non-statistical heightened risk for malignant breast cancer (OR=2.42; 95% CI= 0.1988 to 29.6617; p=0.48: OR=1.66; 95% CI= 0.5270 to 5.2898; p=0.38 and OR=1.55; 95% CI= 0.8660 to 2.794; p=0.13 respectively). Further, the recessive CC genotype was found to be associated with decreased risk of malignant breast cancer in codominant model, though the results were not significant (Table 3). Increased associations were observed between benign breast cancer and heterozygous (TC) and recessive homozygous genotypes (CC) (OR=5.10; 95% CI= 0.4653 to 55.8940; p=0.18 and OR=2.91; 95% CI= 0.8637 to 9.8335; p=0.08 respectively) in codominant model. Also, in dominant model of inheritance, the FGFR2 was found to be nonsignificantly associated with increased risk for benign breast tumor (TC+CC: OR=3.18; 95% CI= 0.9989 to 10.1716; p=0.05). Moreover, a statistically increased risk for benign breast tumor was associated with recessive allele (C) of FGFR2 polymorphism in allele specific model of inheritance (OR=5.44; 95% CI= 2.9735 to 9.9688; p<0.0001).
FGFR2 mRNA Expression and Polymorphism (rs2981582) in Concert with Breast Cancer
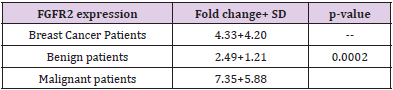
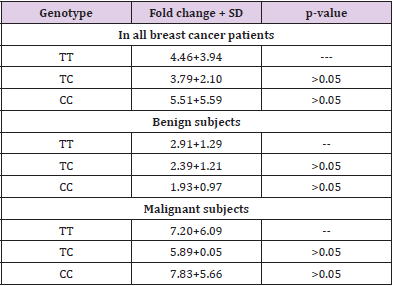
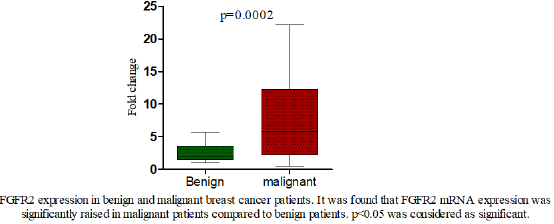
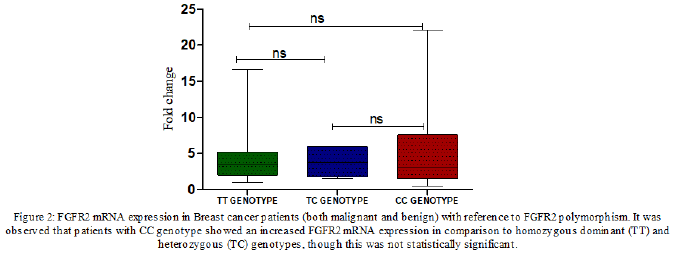
We analysed the mRNA expression of FGFR2 gene and compared it among healthy controls and breast cancer patients. It was observed that RGFR2 mRNA expression varied significantly among healthy controls and breast cancer patients. Breast cancer patients exhibited an increased FGFR2 mRNA expression compared to healthy control subjects (mean fold ± SD=4.33+4.20, Table 3). We further compared FGFR2 mRNA expression among benign and malignant breast cancer patients and observed that FGFR2 mRNA expression was significantly raised by 2.95-fold in malignant subjects compared to benign tumor ones (mean fold ±SD=2.49+1.21 vs 7.35+5.88; p=0.0002 respectively, Table 4 and Figure 1). Next, we categorized patients based on genotypic distribution of FGFR2 (rs2981582) polymorphism and analyzed the FGFR2 mRNA expression in breast cancer patients with reference to their genotype. We observed an increased FGFR2 mRNA expression in patients with mutant homozygous (CC) genotype (mean fold± SD=5.51±5.59) compared to patients with wild type (TT) (mean fold +SD=4.46±3.94) and heterozygous (CC) genotypes (mean fold ±SD=3.79±2.10), but this difference did not reach statistical significance (Table 4 & Figure 2). Similarly, FGFR2 mRNA expression in concert with FGFR2 (rs2981582) polymorphism genotype was analysed in patients of benign and malignant tumour separately (Table 5). However, we could not reach any meaningful inference as the FGFR2 mRNA expression did not follow a meaningful pattern. The results of the analysis are depicted in Table 4 below.
Discussion
In addition to several known risk factors, studies have shown
strong relations with FGFR2 gene in respect to breast cancer.
Multiple genetic aberrations in FGFR2 which triggers the activation
of up and/ or downstream FGFR2 signaling pathways have
been identified in breast cancer [7]. In this study, SNP of FGFR2
rs2981582 of genotype T>C have been chosen for assessing the
association with risk of breast cancer. The protocol resulted in
fragment lengths of 176 base pairs, with genotype patterns wild
type TT [176 base pairs(bp)], heterozygous TC (176+154+22
bp) and homozygous CC (154+22bp).On assessment of specific
genotype association with breast cancer, our results show that
on comparing malignant with healthy controls, a slightly higher
risk is associated with heterozygous genotype TC with OR of
2.4 (95% CI=0.199-29.66) but is not statistically significant (p
value=0.481) TT genotype being the wild type (Table 3). Also,
while comparing malignant with benign cases, no significant risk
in both heterozygous genotype TC (OR=0.47 95% CI=0.067-3.396
P-value=0.46) and homozygous genotype CC (OR=0.53 95%CI =
0.164-1.75 P-value=0.30) (Table 3) was observed. So, rs2981582
SNP did not alter the risk of benign or malignant breast tumour in
this population. A study of 203 histopathologically confirmed cases
of breast cancer and 200 cancer free controls by Liu et.al showed
that the rs2981582 TT genotype is significantly associated with
breast cancer in Chinese women (OR=1.831, p-value=0.037) [18]
but no evidence of significant association was found in this study of 25 breast cancer cases and 25 benign cases with 25 healthy
controls. The small sample size may be attributed to the above
finding in our study.
Our results have shown that FGFR2 expression was significantly
increased in malignant cases (mean fold change =7.35±5.88) as
compared to benign cases (mean fold change=2.49±1.219) with
p-value of 0.0002 (Fig.1). This finding is correlating with the
previous studies on overexpression of FGFR2 in breast cancer
[19]. So we infer that altered expression of FGFR2 might have a
role in pathogenesis/malignant transformation of breast tumor
/ cancer. However, on comparison of FGFR2 expression among
benign patients with different genotypes (rs2981582 T>C) the
differential gene expression associated with different genotypes
of the polymorphism (TT/TC/CC) was not found to be statistically
significant (Figure 2). On comparing the same among malignant
patients, FGFR2 gene expression among different genotypes in
malignant cases was also statistically insignificant (Figure 3). So,
we hypothesize from this study that rs2981582 SNP does not
influence the expression of FGFR2 in this population.
To the best of our knowledge, previous studies have not clearly documented possible relationship between FGFR2 expression with different genotypes of the polymorphism rs2981582. It is worth exploring which SNPs of FGFR2 gene influence its expression or if there is some gene-gene or gene-environment interaction influencing its expression in breast cancer.
Declarations
a) The authors declare that there are no competing conflicts of
interest
b) No funding was received from any agency for conducting this
study
c) All the details are mentioned in the manuscript and no
supplementary files are available. All the data is available.
d) Ethics approval was obtained from the institutional ethics
committee
e) All the participants were recruited after their written consent
f) All the authors have provided their consent for publication of
the study findings.
References
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, et al. (2015) Global cancer statistics, 2012. CA Cancer J Clin 65: 87-108.
- Martinez MT, Oltra SS, Chilet MP, Alonso E, Hernando C, et al. (2019) Breast cancer in very young patients in a Spanish cohort: Age as an independent bad prognostic indicator. Breast Cancer: Basic and Clinical Research 13:1-10.
- Lee YC, Chuang JP, Hsieh PC, Chiou MJ, Li CY (2015) A higher incidence rate of acute coronary syndrome following radiation therapy in patients with breast cancer and a history of coronary artery diseases. Breast Cancer Res Treat 152: 429-435.
- Huang Y, Song F, Chen K (2015) Current status of genome-wide association studies (GWAS) on breast cancer and application values of single nucleotide polymorphisms identified from GWAS. Zhonghua Liu Xing Bing Xue Za Zhi 36: 1058-1061.
- Raskin L, Pinchev M, Arad C, Lejbkowicz F, Tamir A, et al. (2008) FGFR2 is a breast cancer susceptibility gene in Jewish and Arab Israeli populations. Cancer Epidemiol Bio Prev 17: 1060-1065.
- Liang J, Chen P, Hu Z, Zhou X, Chen L, et al. (2008) Genetic variants in fibroblast growth factor receptor 2 (FGFR2) contribute to susceptibility of breast cancer in Chinese women. Carcinogenesis 29: 2341-2346.
- Lei H, Deng CX (2017) Fibroblast Growth factor Receptor 2 Signaling in Breast Cancer. Int J Biol Sci 13(9): 1163-1171.
- Touat M, Ileana E, Postel-Vinay S, Andre F, Soria JC (2015) Targeting FGFR Signaling in Cancer. Clin Can Res 21: 2684-2694.
- Turner N, Grose R (2010) Fibroblast growth factor signalling: from development to cancer. Nat Rev Can 10: 116-129.
- Sun S, Jiang Y, Zhang G, Song H, Zhang X, et al. (2012) Increased expression of fibroblastic growth factor receptor 2 is correlated with poor prognosis in patients with breast cancer. J Surg Oncol 105: 773-779.
- Wesche J, Haglund K, Haugsten EM (2011) Fibroblast growth factors and their receptors in cancer. Biochem J 437: 199-213.
- Easton DF, Pooley KA, Dunning AM, Pharoah PDP, Thompson D, et al. (2007) Genome-wide association study identifies novel breast cancer susceptibility loci. Nature 447: 1087-1097.
- Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, et al. (2007) A genome- wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet 39: 870-874.
- Andre F, Cortes J (2015) Rationale for targeting fibroblast growth factor receptor signaling in breast cancer. Breast Cancer Res Treat 150: 1-8.
- Katoh M (2019) Fibroblast growth factor receptors as treatment targets in clinical oncology. Nat Rev Clin Oncol 16: 105-222.
- Byron SA, Pollock PM (2009) FGFR2 as a molecular target in endometrial cancer. Future Oncol 5(1): 27-32.
- Wang Y, Zhang H, Wang Y (2018) Relevance between FGFR2 gene polymorphism and breast cancer. Acta Medica Mediterrania 34: 1717.
- Liu CL, Hu XP, Guo WD, Yang L, Dang J, et al. (2013) Case-Control Study on the Fibroblast Growth Factor Receptor 2 Gene Polymorphisms Associated with Breast Cancer in Chinese Han Women. J Breast Cancer 16(4): 366-371.
- Sun S, Jiang Y, Zhang G, Song H, Zhang X, et al. (2012) Increased expression of fibroblastic growth factor receptor 2 is correlated with poor prognosis in patients with breast cancer. J Surg Oncol 105: 773-779.

 Research Article
Research Article