Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Arash Kashani1, Benjamin William Behrens Holman2 and Aduli Enoch Othniel Malau-Aduli1,3*
Received: July 17, 2017; Published: July 24, 2017
Corresponding author:Aduli Enoch Othniel Malau-Aduli, Veterinary and Biomedical Sciences, College of Public Health, Medical and Veterinary Sciences, Division of Tropical Health and Medicine, James Cook University, Townsville, Queensland 4811, Australia
DOI: 10.26717/BJSTR.2017.01.000218
The adrenergic-receptor beta3 (ADRB3) gene is an obesity gene that is involved in the regulation of energy balance and a variety of physiological functions by increasing lipolysis and thermogenesis. Spirulina (Arthrospira platensis) is a blue-green cyanobacterial alga containing 60-70% protein with an extensive history of human consumption, and more recently, inclusion in animal feeds. We had earlier demonstrated that low level oral supplementation by drenching prime lambs with 100 mL/head/day of 1g of Spirulina powder dissolved in 10 mL of water (10%wt:vol) increased live weight and body conformation measurements in Black Suffolk (BS) x Merino crossbred sheep. The hypothesis that genetics-nutrition interactions between sheep breeds with fewer mutations at the ADRB3 locus and an optimal Spirulina supplementation level will increase lean meat production was tested in the current study. Forty-eight crossbred Australian prime lambs sired by four rams of diverse breeds under the same pasture-based management conditions were subjected to a nine-week feeding trial with Spirulina, followed by genomic DNA extraction and single nucleotide polymorphism (SNP) analysis. Eleven SNPs in both the coding and noncoding regions of the ovine ADRB3 gene were detected. Nine of the SNPs were in exon I and two in the intron. Variations in SNP frequencies were highly significant (P <0.0001) between all sheep breeds. The maximum and minimum number of SNPs were found in purebred Merinos (4.83) and Black Suffolk x Merino (BS) crossbreds (1.67). In total, one indel and six transverse mutations were detected that resulted in six amino acid substitutions. BS crossbreds had the lowest frequency of mutation and amino acid substitutions in their population in agreement with our hypothesis. In conclusion, BS sheep genetics matched with low level (100 mL/head/day) of Spirulina supplementation can lead to higher meat production with less fat content in a typical pasture-based sheep production system.
Keywords:Cerebral Palsy; Parents; Children; Adolescents; Quality Of Life; Physiotherapy
Sheep are one of the most economically important species of domesticated livestock for producing milk, meat, skin, and wool for humans [1,2]. In meat production, saleable meat yield is a crucial determinant of the financial returns and economic value accruable to producers. It is estimated by the ratio of muscle to carcass weights [3]. Fat content in the carcass is an essential element for the perception of texture, flavor and juiciness. It is also a source of essential fatty acids that cannot be synthesized by humans, but nowadays, it is considered as an unpopular and unhealthy meat constituent by many consumers [4]. Hence, researchers have been keenly interested in exploring ways and means of manipulating the fatty acid composition of meat by altering the ratio of saturated (SFA) to unsaturated (USFA) fatty acids. Achievement of this feat seems possible with the use of molecular markers in animal breeding program in combination with strategic nutritional supplementation with nutrient-dense feeds. Therefore, existing knowledge gaps in genetics-nutrition interactions for optimal lamb performance and healthy meat production need to be filled. This study intends to contribute in this regard utilizing Australian prime lambs and dual-purpose sheep production system.
The β3-adrenergic receptor (ADRB3), also known as β3- adrenoreceptor, is a G-protein coupled receptor predominantly located in the adipose tissue [1,5]. It plays a major role in regulating mammalian energy storage and expenditure under mediating effects of the sympathetic nervous system [5,6]. It is also the main mediator of the lipolytic and thermogenetic effect of high catecholamine (in particular norepinephrine) concentration in brown and white adipose tissues in rodents [6,7]. The receptor’s primary role is speculated to be in regulating resting metabolic rate and lipolysis [8]. In humans, polymorphisms in the ADRB3 gene have been associated with diabetes and obesity, where tryptophan is replaced by arginine at position (Try64Arg) [1,5]. In obese rats, a decreased level of ADRB3 expression has been found in brown and white adipose tissue [5]. In addition, mice with a disrupted ADRB3 gene had a decrease in lean body mass and a modest increase in body fat [5]. In sheep, polymorphism of the ADRB3 gene has been associated with the economically important quantitative traits of birth weight, growth rate, carcass composition, cold survival, wool staple strength and wool yield [5,7,8].
Spirulina (Arthrospira platensis), an edible blue-green microalga, is a human and animal nutritional supplement that contains about 60%-70% protein, all essential amino acids, carotenoids, vitamins, and minerals [9-12]. Kulpys et al. [13] demonstrated that dairy cows supplemented with Spirulina had a 21% increase in milk production. Holman & Malau-Aduli [12] have published a review of the importance, sustainability and commercial production of Spirulina with the global locations of production companies in Australia and other countries around the world. In dual-purpose prime lamb production, profitability is driven primarily by protein-rich supplementation. As a result, a gain in higher lamb production with healthier meat composition is a subject of immense interest, particularly in the highly successful Australian sheep industry. However, to our current knowledge, there is no published investigation of polymorphisms of the ovine ADRB3 gene and its association with production traits utilizing Spirulina supplementation strategy anywhere in the world. Therefore, our research objective was to detect polymorphisms of this gene in Australian crossbred sheep and evaluate its association with Spirulina supplementation level with a view to potentially increasing lamb production with low fat content.
Animals, management and phenotypic data collection. All procedures involving animals were approved by the University of Tasmania Animal Ethics Committee, and were conducted in accordance with the 1993 Tasmanian Animal Welfare Act and the 2004 Australian Code of Practice for the Care and Use of Animals for Scientific Purposes. The experimental flock at the University of Tasmania Farm, Cambridge, Hobart, utilized for this study comprised forty-eight weaned lambs from purebred Merino dams sired by White Suffolk, Black
104 Suffolk, Dorset, and Merino rams under the same management conditions. All animals were maintained on ryegrass pastures as the basal diet. At six weeks of age, they were balanced by sire breed and gender and randomly allocated into three treatment groups: the control group grazing without Spirulina (0%), low (100mL/head/day in the ratio of 1g of Spirulina powder: 10mL of water or 10% wt/vol), and high (200mL/head/day in the ratio of 2g of Spirulina powder: 10mL of water or 20% wt/vol) Spirulina supplementation levels. The Spirulina powder was purchased from a commercial producer in Darwin, Northern Territory, Australia (TAAU, NT, Aus). Lambs were daily supplemented according to their assigned Spirulina treatment before being released into paddocks for grazing. The supplementary feeding trial continued for nine weeks after an initial three weeks of adjustment.
Liveweight (LWT), body condition score (BCS), body length (BL), withers height (WH), chest girth (CG), and average daily gain (ADG) were recorded weekly over this period. Details of the procedures for recording these growth and body conformation measurements had previously been described [11]. At the end of the feeding trial, blood samples were taken by jugular venipuncture before the animals were slaughtered at a commercial abattoir in Gretna, Tasmania, for carcass and sensory evaluation of meat quality.
Blood samples, after collection from the forty eight weaners, were stored at -80 °C until ready for genomic DNA extraction. Genomic DNA was extracted from the samples in triplicates (total of 144 samples) using Ultraclean Tissue and Blood Spin DNA Isolation Kits (MoBio, Solana Beach, CA). The purity of the extracted DNA was quantified using the NanoDrop 8000 spectrophotometer (NanoDrop, Wilmington, DE).
To amplify a series of overlapping fragments of the ovine ADRB3 gene sequence (Gene Bank Accession Number DQ269497) [1], seven pairs of PCR and interrogation primers were designed using Primo SNP 3.4: SNP PCR Primer Design software (Chang Bioscience 2004). The PCR fragments were amplified from 20 ng of genomic DNA in a total volume of 20 μL with 10 μM of each dNTP, 2 mM MgCl2, PCR primers in various concentrations (10-25 fM/μL), and 0.5 U μL HotStartTaq DNA polymerase (Qiagen, Inc.). The PCR cycling profile was: initial denaturation at 95ºC for 10 min followed by 35 cycles of denaturation at 94ºC for 30 s, primer annealing at 64ºC for 30s, and elongation at 72ºC for 1 min. Final extension was at 72ºC for 3 min. To remove any remaining single-stranded primers and dNTPs, 2 μL of the PCR products was treated with 5 μL Antarctic phosphatase-exonuclease I buffer at 37ºC for 15 min, then incubated with 7 μL Antarctic phosphatase buffer at 37ºC for 15 min, and finally heat-treated at 80ºC for 20 min. The PCR products were electrophoreses on agarose gels to verify specificity and to rule out any artefact bands.
The Genome LabTM SNP Start Primer Extension Kit (PN A23201) was employed for SNP assay according to the manufacturer’s instructions. To 4 μL of SNPStart Master Mix, 1 μL of interrogation primer, various concentrations of PCR products (100 fM), and DNAse and RNAse-free PCR grade water were added, creating a 10 μL final volume. The recommended two-step protocol was: 90ºC for 10s followed by 45ºC for 20s, cycled 35 times. To remove any remaining unincorporated dye-labelled terminators, a simple digestion was performed using shrimp alkaline phosphate (SAP). About 0.25 U SAP, 1 μL SAP buffer, and 1.75 μL DNAse- and RNAse-free PCR grade water were used, and the samples incubated for 30 min at 37ºC followed by 15 min at 65ºC to inactivate the SAP enzyme. Then 1 μL of the purified SNP reaction, 0.5 μL size standard 80 (PN608395), and 39.5 μL sample loading solution (SLS) for each sample were mixed and loaded on a fresh sample plate (PN609801) and overlaid with mineral oil. The sample plate was loaded onto the GenomeLab/CEQ Genetic Analysis System Sequencer. Sequencing was performed using the Genome Lab TM Sequencing Kit (PN608000) procedures.
Alleles were separated and detected using the Genome Lab TM SNP Start Primer Extension Kit and SNP locus tags via capillary electrophoresis on the Beckman Coulter CEQTM 8000 Series Genetic Analysis System. The software SNP Analysis Parameters was employed to analyze the data generated. Apparent fragment size and allele ID options were added and the samples on an ABI 3100 genetic analyzer (Applied Biosystems). Further processing by Genemapper software (Applied Biosystems) and multiple sequence alignments were carried out using DNAMAN (version 6.0, Lynnon Biosoft, USA) to confirm allele separation and SNP genotyping. Genotype frequencies were tested for deviations from the Hardy- Weinberg equilibrium using χ165 2 tests.
The frequency of each SNP was coded as present (1) or absent (0) for each animal’s genotype and threshold non-linear model procedures in SAS (SAS Inst., NC) used to analyze the data. Then a generalized linear model (PROC GLM, SAS Inst., NC) was used to fit the fixed effects of SNP, sire breed, sex, supplementation level, and their interactions on SNP frequency distributions, growth, and body conformation parameters. Significance levels were tested at p<0.01 and p<0.05 using Tukey pair wise comparison thresholds.
The PCR and interrogation primers designed for amplifying the ovine ADRB3 gene worked well on all blood samples under the optimized platform.
As depicted Spirulina supplementation enabled sheep to have greater BL than the control group (p < 0.015). Furthermore, lambs in the high (200mL/head/day of 20%wt/vol) Spirulina supplementation treatment group had greater BCS than the low (100mL/head/day of 10%wt/vol) and 0% (control) treatment groups (p < 0.001). It was observed that sheep receiving low Spirulina supplementation had the heaviest LWT, 41.9 kg (p < 0.018); however, no difference was found between the high and control groups.
Three unique patterns were detected using 7 primer sets. All loci were heterozygous genotypes. The frequencies for alleles A, B, and C were 6.1%, 85.35%, and 8.49%. Allele B was present in all breeds, whereas allele A was found in White Suffolk, Dorset, and Merinos and allele C was present in White Suffolk and Merinos only.
Fragment analysis and sequence comparison revealed that variations in the ovine ADRB3 gene occurred in both the coding and non-coding regions. In total, 11 SNPs were identified, of which 9 were present in the exon and the other two in the intron. From these eleven SNPs, one indel, two transitions, six transversions in exon I, and one transition in the intron were found. In addition, the remaining SNP belonged to the three-base substitution class (1733 A>G>C) which was found in the intron that could have been either a transition or transversion. The six transversions in exon I changed the amino acid composition.
Differences existed between breeds and sexes and the GLM test showed that the differences were highly significant in breeds (p < 0.0001) and sexes (p < 0.0081). Purebred Merino sheep had the maximum average SNP number (4.83) and Black Suffolk crossbreds had the minimum (1.67). The average SNP numbers in White Suffolk and Dorset crossbreds were 3.0 and 4.17, respectively. Tukey’s pairwise mean comparison test indicated that these significant breed differences resulted mainly from all breeds. In addition, the number of SNPs was higher in wethers (3.58) than ewes (3.25).
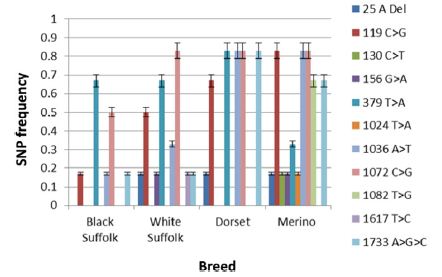
Figure 1: SNP frequencies in exon 1 and intron 1 by breed of sheep.
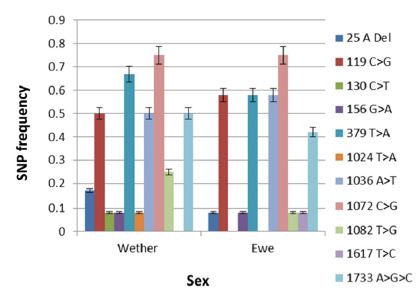
Figure 2: SNP frequencies in exon 1 and intron in 17 wethers and ewes.
The frequency of SNPs in the various breeds and sexes (exon I) are shown in (Figures 1 & 2), respectively. SNP1 (25 A Del) occurred with significantly higher (p < 0.0001) frequency in White Suffolk (WS), Dorset (D) and Merino (M) breeds than Black Suffolk (BS). In addition, SNP1 was expressed with significantly higher frequency (p < 0.0015) in wethers (0.15) than in ewes (0.08). The 119 C>G (SNP 2) presented in all breeds as a missense mutation which caused an Ala40Gly mutation. The maximum frequency of SNP 2 was found to be significantly higher (p < 0.0001) in M (0.83) than in D (0.67), WS (0.50), or BS (0.17). It had a slightly higher frequency in ewes (0.58) than in wethers (0.50).
The frequency of SNP3 was greatest among M sheep (0.17) and this breed difference was highly significant (p < 0.0001). The 130 C>T (SNP3) was a transition type mutation which did not occur in ewes, although it had a significant frequency (p < 0.0001) in wethers (0.08). Similarly, SNP4 (156 >A) was a transition type mutation, but was found in both M (0.17) and WS (0.17) breeds with significant breed differences in frequencies (p < 0.0001). It presented equally in both wethers (0.80) and ewes (0.80), thus no significant difference was observed. A missense mutation, SNP5 (379 T>A), resulted in a Ser127Thr mutation. It significantly (p < 0.0001) differed in all breeds (D, 0.83; both BS and WS. 0.67; and M, 0.33) and was significantly higher (p < 0.0194) in wethers (0.67) than ewes (0.58). SNP6 (1024 T>A) was found exclusively in M sheep (0.17) and significantly differed (p < 0.0001) from other breeds. It was a transversion mutation which caused a missense mutation of Tyr342Asn. Its frequency was observed only in wethers (0.08) and was significantly different (p < 0.0001) from ewes. The 1036 A>T (SNP7) was discovered in all breeds with significantly higher (p < 0.0001) frequencies in both D and M (0.83), WS (0.33), and BS (0.17). Its frequency was observed to 236 be significantly (p <237 0.0017) higher in ewes (0.58) than wethers (0.50).
SNP8 was detected in all breeds, but occurred at a high frequency in WS, D, and M (0.83 each), and a lower frequency in BS (0.50). The 1072 C>G (SNP8) mutation was a missense Arg358Gly mutation. Its frequency significantly differed (p <0.0001) in all breeds. There was no significant sex difference between lambs. Another missense mutation, 1082 T>G (SNP9), resulting in Phe361Cys, was detected exclusively in M (0.67) breed. Its frequency significantly differed (p < 0.0001) between breeds and sexes; being higher in wethers (0.25) than ewes (0.08). In the intron, SNP10 (1617 T>C) was a transition mutation that mainly presented in WS (0.17) and significantly differed from other breeds (p < 0.0001). It was detected only in ewes (0.08) and significantly differed from wethers (p < 0.0001). Finally, the 1733 A>G>C (SNP11) is a tri-base mutation seen in all breeds, but with significantly higher (p < 0.0001) frequency in D (0.83), M (0.67), WS (0.17), and BS (0.17). Its frequency was significantly (p < 0.0291) higher in wethers (0.50) than ewes (0.42).
This experiment is a follow-up to our previous results demonstrating that the live weight and body conformation measurements of Australian crossbred sheep can be boosted by using protein-rich Spirulina as a dietary supplement [11]. In agreement with the previous data, demonstrated that at low level of Spirulina supplementation, there was an increase in live weight and body conformation traits. However, excessive Spirulina supplementation can suppress optimal sheep growth because the excess protein is deaminated and gets lost in the urine or broken down in the liver, leading to fatty liver and ketosis [14]. In addition, we confirmed the importance of sire breed selection in breeding programmes because of variations in genetic predisposition for muscle growth as opposed to body fat deposition [15].
The current study describes polymorphisms (SNP variation) in both coding and non-coding regions of the ovine ADRB3 gene in purebred and crossbred Australian sheep and its association with Spirulina supplementation. Indel mutations play key roles in the ovine genome and can result in frame shift mutations of the amino acid sequence. However, a comprehensive understanding of how the indels influence the phenotype and impact evolutionary processes is not clearly understood [1]. SNP2 (119 C>G) mutation resulted in Ala40Gly substitution which is a change from the hydrophobic alanine (A) to glycine (G) at position 40. SNP5 (379T>A) mutation is responsible for Ser127Thr substitution. This substitution is a change from the small sized and polar Serine (S) to the medium sized and polar Threonine (T) at position 127. These two substitutions are located in the first and third transmembrane segments in the primary structure of the ovine ADRB3 locus, respectively, and have residues common to all two β-adrenergic receptor subtypes [1, 16]. These amino acid substitutions are thought to be involved in ligand binding [1,16]. The finding that SNP2 was distributed at 275 a higher frequency in purebred Merino (M) than in other breeds, and SNP5 distribution was higher in Dorset (D) crossbreds, is consistent with a recently published study that reported higher frequencies of these substitutions in dual purpose meat and wool sheep breeds [1].
Both SNP6 and SNP7 responsible for Tyr342Asn and Asn346Tyr substitutions respectively, are located in the seventh transmembrane segment of the ovine ADRB3 gene [16,17]. It has been shown that some residues in this segment are vital for receptor activation by ADRB3 agonists [18] and for ligand binding [16]. Both substitutions Arg358Gly (SNP8) and Phe361Cys (SNP9) are located in the carboxyl terminal region of the ADRB3 receptor. This region plays a crucial role in cell signaling efficiency [17] and houses the receptor sites for coupling with G-protein [16]. These Arg358Gly and Phe361Cys substitutions can result in change or loss of function of the ADRB3 receptor. It has been demonstrated that the oral administration of β-adrenergic agonists increased muscle accretion and decreased fat deposition in pigs and cattle [19]; thus, the functional change can affect meat production and quality. The mutation frequency of SNP8 was low in Black Suffolk (BS) crossbreds and high in White Suffolk (WS) and D crossbreds, and purebred M lambs. The Phe361Cys (SNP9) frequency was detected only in M lambs. In the intron, SNP10 was detected in WS crossbreds only, while SNP3 was only detected in M purebreds, suggesting the use of these SNPs as molecular markers for breeding
programs. The total SNP frequencies significantly differed between lamb breeds; reinforcing the underlying fact that polymorphisms in the ADRB3 gene can affect the expression, tissue distribution, thermogenic and lipolytic capabilities of the receptor [7,20,21].
There is a significant association between Trp64Arg mutation in human ADRB3 gene with obesity [22-24], insulin resistance and hyperuricemia [25,26]. The associations between ovine ADRB3 polymorphism with birth weight, growth rate, carcass composition, lamb mortality and cold survival and wool mean staple strength and yield have been confirmed [7,8,21]. This current study established the existence of a significant SNP difference between breeds of Australian purebred and crossbred sheep and their importance in changing the ADRB3 gene function. In this study, we detected less mutations in the BS breed compared to other breeds, particularly D crossbreds and M purebreds. These mutations result in amino acid changes which may increase fat deposition in sheep tissues. Thus, in conjunction with our previous findings from the Spirulina supplementation experiment, we suggest the use of a combination of BS breed and low Spirulina supplementation in order to increase lamb production with less fat content in the meat. However, further fatty acid profiling, mRNA expression and proteomic analyses are required to fully understand the underlying mechanisms of ADRB3 SNPs and gene expression in response to Spirulina supplementation.
The ovine ADRB3 gene was investigated, several SNPs identified and their association with Spirulina supplementation level evaluated in purebred and crossbred lambs. The identified SNPs in both coding and non-coding regions and their distributions in four sheep breeds reflected existing 314 genetic variation. In conclusion, the genetics-nutrition combination of supplementing Black Suffolk crossbred lambs with 100mL/head/day of 10%wt/vol of Spirulina (low level of supplementation) may be used to increase lean meat production in the Australian prime lamb industry. Therefore, the hypothesis that genetics-nutrition interactions between sheep breeds with fewer mutations at the ADRB3 locus and an optimal Spirulina supplementation level will increase lean meat production holds true and should be accepted.
This research was funded by grants and postgraduate scholarships from the University of Tasmania (UTAS) and the Australian Wool Education Trust (AWET). We thank Chris Gunn, John Otto, Will Bignell and Barrie Wells for their inputs during the sheep breeding and feeding trials.


