Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Jean-Claude Perez *
Received: September 07, 2017; Published: September 20, 2017
Corresponding author: Jean Claude Perez, £7 avenue de Terre-rouge F33127 Martignas Bordeaux metro pole, France
DOI: 10.26717/BJSTR.2017.01.000369
For more than 25 years, we have been looking for biomathematical laws controlling and structuring DNA, genes, proteins, chromosomes and genomes [1,2]. In 1997 we discovered a simple numerical law based solely on atomic masses, unifying the 3 languages of biology: DNA, RNA, and amino acids. This law, “The Master Code Of Biology” is published in 2009 in the book codex biogenesis [3] and then in 2015 in a reference article peer review [4]. In 2017 we publish different applications: HIV, SNPs, brain genetics [5-8]. On the other hand, at the beginning of the 2000s we studied the integrity of the Prions proteins (CJD, Creutzfeldt-Jakob disease), showing a common invariant bi-attractors in “W”, while the amyloid was characterized by a pattern in “V” shape. Since that time, biologists such as Claudio Soto have visually demonstrated in 3D the same biological form in horseshoe for the amyloid protein [9-13].
Keywords: Biomathematics; Master Code; DNA; Prions diseases; Genomics; Proteomics
Figure 1: Master Code Genomics/Proteomics pattern analysis demonstrates evidence of 2 attractor
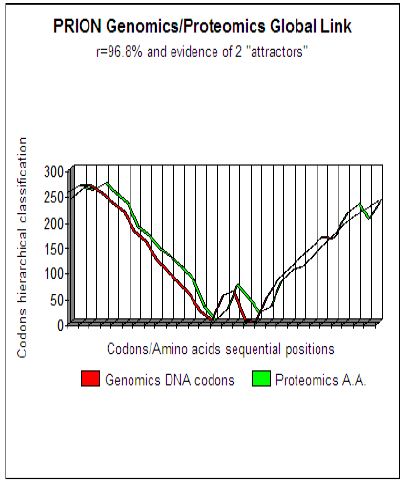
Concretely, the dynamics of this Genomics/Proteomics coupling is appeared as two correlated curves translating topology and the dynamic evolution of a hierarchical classification of codons throughout the studied sequence. More precisely, the study of these curves shows that this tool highlights the functional areas and active sites within proteins. The specific context of Prions area could be an interesting field to run and improve our new basic research approach. Studying Prions by this method underlines two remarkable facts:
a) A quasi-perfect coupling between genomics and proteomics sequences (about 96 to 98% for the most common Prions).
b) Two concurrent, distant sites of approximately 40 amino acids are underlined. We thought that these two concurrent sites could constitute two “functional attractors “ which could be at the origin of the two stable structures PrPc and PrPsc of Prisons (Figure 1).
We thus sought to test, simulate and validate this thesis according to the four following protocols of experiments:
Unification of the 167 Prions of GenBank highlighting of an Invariant: We analyzed systematically all GenBank available Prions and Prions-Like. The second of the two sites presented above (Major site towards codon 138 in human Prion) appears in the majority GenBank Prions in the form of an optimum consensus centered on the area rpIihfgs This Unification is very beyond the “barrier of the species “, the lengths of Prions studied and even of the precise values of each amino acids. For information, more than 83% of the 167 Prions studied is unified on this “pattern”. In other hands, more than 3 Prions by 4 have a Genomics/Proteomics coupling higher than 95%.
Thesis of self-assemblies PRION PRION or PRION Scrapie following a shift around 40 codons: We imagined various scenarios of interactions Prions Prions articulated around these 2 sites. One of them leads to a perfect coupling of double interactions DNA/RNA strains and Proteomics/Proteomics strains. There is about a self-assembly between both Prions proteins with a relative shifting by approximately 40 codons (which constitutes also the amino acids distance between the 2 sites). We test these hybrid assemblies for the man, the cow, the sheep, the mouse and the rabbit. One distinguishes 2 cases PRION Prion and Prion Scrapie (Golden Hamster). Unification Genomics/Proteomics is always >92% on average. This possible interaction is “above “ the concept of species. The optimal shift varies from 37 to 40 codons according to species.
Validation of our thesis on various experiments linked with pathogenicity (sheep, human cjd, “prusiner miniprions “, etc): Then, we run our thesis on experimental published cases connecting changes and pathogenicity of Prions. Thus, we show that the quality of the Genomics/Proteomics coupling for the 40 codons shift based self-assembly moves in the same direction as the sheep pathogenicity (mutations sets VRQ, ARH, ARQ, ARR and AHQ). In the same way, we associate coupling Genomics/Proteomics and pathogenicity for the man (changes 102, 129/178 and GSS/FFI/ CJD). Lastly, our model completely confirms the experiments of Professor Stanley prusiner on the “Miniprions “.
Extension of our Assumption to the field of the “Prions-Like” (URE3, SUP35P Yeast and Homologues): Can the Invariants and self-assembly thesis be wide until the related field of “Prions-Like”? Then, we studied URE3: it appears a pattern similar to Prions ones (double attractors), moreover we discover quickly a possible selfassembly URE3 URE3 (optimal shift of 15 codons and coupling r=91.7%). In the same way a generalization is running on SUP35 yeast and homologues. For example, in the Drosophila sample (GenBank: U88868), Dsup35 Prion-like produces a dynamics of Genomics/Proteomics curve similar to the above regular Prions curves. A self-assembly shift thesis is also proposed.
It is a “new point of view” of the Prions field which are proposed here. Beyond a support “Bio-Mathematics”, we UNIFY information Genomics/Proteomics of all them Prions. We propose and validate a thesis, plausible, of self-interaction between Prions. This model is beyond and apart from the “barrier of the species”. Our thesis seems confirmed by multiple experiments published; it would seem strongly associated with the pathogenicity. Lastly, our model of interaction Prion Prion seems to extend to the “Prions-Like” and even to other peptides: we validated it, therefore [12], on the formation of Amyloids plaques (Beta A4 Amyloid ALZHEIMER, particularly peptide A-beta 1-42).
As demonstrated by PR claudio Soto in [10] “Alzheimer’s diseases transmission may be similar to infectious prion diseases”. Then particularly the Master Code Of Biology biomathematical approach révélas also - strong relationships between our simulations on Prions and on Amyloids self-assemblies. Both Prion and most probably Amyloid are proteins whose dynamics interactions are beyond the state of the art of conventional Biology. Therefore, we believe that multidisciplinary approaches close to quantum physics may allow us to understand how they work [13].


