Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Shadia AH Fathy1, Manal A Emam1, Sara HA Agwa2, Christina A.Soliman1 and Sarah El-Nakeep*3
Received: January 02, 2018; Published: January 11, 2018
*Corresponding author: Sarah El-Nakeep, Hepatology and Gastroenterology Unit,Internal Medicine Department, Faculty of Medicine, Ain Shams University, Cairo, Egypt
DOI: 10.26717/BJSTR.2018.02.000652
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the leading cause of cancer death in the world. Its poor prognosis is mainly due to the late detection of the tumor, thus multiple genetic markers including micro RNAs are under research to improve detection and prognosis of HCC.
Aim: of this study was to evaluate the diagnostic potential of a panel of two circulating miR 143 and miR-550a as diagnostic biomarkers for hepatocellular carcinoma (HCC) among patients with hepatitis C virus (HCV) infected Egyptian.
Methods: A total of 81 participants were included in this study (3 controls, 41 hepatitis C patients and 37 HCC patients). Samples were collected, and both miR143 and miR-550a were assessed.
Results: The expression levels of the two miRNAs (miR-143 and miR-550a) were detected by quantitative reverse-transcriptase PCR (RTqPCR) in controls, HCV and HCC patients. miR-143 showed no correlation with the two groups while miR-550 a showed reversed correlation. AUC for miR550 was 0.93 (0.86-0.99) with sensitivity 91.89% and specificity 90.24% at the cutoff value ≤3.29 for prediction of HCC occurrence, and NPV was 92.5 and PPV was 89.5.
Conclusion: The expression of plasma miR-143 was not correlated with the occurrence of HCC while plasma miR-550a was significantly down-regulated in patients with HCC, thus it could be a good potential circulating biomarker for HCC.
Keywords: Hepatocellular carcinoma; hepatitis C virus; miR-143; miR-550a; HCC prognosis; HCC diagnosis
Abbreviations: HCC: Hepatocellular Carcinoma; CT: Computed Tomography; MRI: Magnetic Resonance Imaging; ALT: Alanine Aminotransferase; AST: Aspartate Amino-Transferase; INR: International Normalized Ratio; AFP: Alpha-Feto Protein; SPSS: Statistical Program for Social Science; AUC: Area Under Curve; ROC: Receiver Operating Characteristic
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer [1]. Overall, it is the fifth most common cancer worldwide, representing about 5.7% of all new human cancer cases with an increasing incidence expected in the next decade [2]. High prevalence of HCC in Egypt is mainly caused by the high prevalence in HCV; where an Egyptian cohort of HCC patients found that the prevalence of HCV was 91.32% while HBV was 2.51% in the studied population [3]. HCV related HCC is preceded by cirrhosis, in contrary to HBV where HCC is induced by the genetic mutation caused by the virus rather than its liver destruction effect by chronic hepatitis, thus in HBV HCC could present earlier that HCV [4]. The ASLD and EASL guidelines recently removed AFP from the screening of HCC as its diagnostic efficacy is high only when there is micro-metastasis or late stage of cancer, a compelling need for early detection markers for better prognosis of HCC and rendering the patient more liable to curative measures, to improve the patients survival [5].
Curative measures for HCC patients are still limited to the early discovery of the disease and includes; liver transplantation, resection or radiofrequency [6]. MicroRNAs (miRNAs) are endogenous, small, single-stranded, noncoding RNAs that consist of 20 to 25 bases. They control various biological functions such as cellular proliferation, differentiation and apoptosis. Approximately half of human miRNAs are located in fragile regions chromosomes, which are associated with the development of human cancers. miRNAs have been referred to as so-called oncomirs for their functions as tumor suppressors and oncogenes [6]. Aberrant expression of miRNA has been linked to a variety of cancers, including HCC [7]. In the present study, we have evaluated the possibility of using two circular HCC-related miRNAs (miR-143 and miR-550a) as possible non-invasive serum biomarkers for diagnosis and prognosis of HCV associated HCC using qT-PCR. To our knowledge, this is the first study testing mi-143 and miR-550a in plasma of for HCC, also the first to be done in Egypt.
The study population consisted of 41 patients with Chronic Hepatitis C virus and 37 patients with HCV associated hepatocellular carcinoma who were treated at the Tropical Medicine Department and Internal Medicine Department, AinShams University Hospitals and 3 healthy normal volunteers with matching age and sex to the patients’ groups after obtaining informed consent. The study was approved by AinShams Faculty of Medicine Ethical Committee. All of the HCC patients were diagnosed by triphasic contrast enhanced computed tomography (CT) scan or magnetic resonance imaging (MRI), and tested for liver functions and AFP. HCC patients with no evidence of local invasion or distant metastasis and positive for HCV were included. While patients with metastatic HCC or other forms of cancer other than HCC were excluded. Three milliliters of peripheral blood were collected from the patients and healthy controls; plasma was separated for miRNAs purification. All the samples were stored at -80 0C for extraction of RNA.
Laboratory investigations included; liver function tests, alanine aminotransferase (ALT), Aspartate amino-Transferase (AST), total bilirubin, albumin, platelets count and International normalized ratio (INR) were done for all of the subjects in the study. Data of all subjects were obtained from medical records and personal history and clinical examination.
Sera from chronic HCV and HCC patients were used for estimation of serum level of Alpha-feto protein (AFP) by solid phase two sequential chemiluminescent immunometric assay using immulite 2000 system analyzer, the kits were supplied by Siemens (Siemens Healthcare Diagnostics, United States, cat#L2KAP2).
Total RNA, including microRNA was isolated from HCC, HCV and control plasma samples using a mirVana™ PARISTM isolation kit (Ambion, Part Number AM1556) following the enrichment procedure for small RNAs purification according to the manufacturer’s protocol [8]. Control samples were used to obtain the baseline for the genetic markers where controls represented by one fold. The concentration and purity of RNA were determined by measuring the absorbance at 260 and 280nm using NanoDrop 2000A Spectrophotometer (ThermoScientific, USA).
The expressions of miRNAs were quantified according to the manufacturer’s instructions by TaqMan miRNA assays (Applied Biosystems) following reverse transcription (Tiangen, Chin) of 40 ng RNA. Reactions were loaded onto a 96-well plate and run in duplicate on an ABI 7900 Fast Real-Time PCR System (Applied Biosystems). The reactions were firstly incubated at 50°C for 20 seconds and then 95°C for 15 minutes, followed by 40 cycles of denaturation at 94 °C for 15 seconds, then 30 seconds of annealing/extension at 55°C and finally 70 °C for 30 seconds. The ΔΔCT method was used to determine the relative expression level (fold change) for miRNA143 and microRNA 550a [9]. The U6 was chosen as the endogenous normalizer. The primers used for qRT-PCR in this study are as following: miR-143: GCTGAGATGAAGCACTGAAGCTC, miR-550a: AGUGC CUGAGGGAGUAAGAGCCC and U6: CAAAGTCAGTGCAGG TAGGCTTA.
The statistical analyses in this study were performed using the Statistical Program for Social Sciences (SPSS) software 22.0 (SPSS Inc., Chicago, USA). Data were summarized as a mean ± standard deviation. When P<0.05, the difference was considered to have statistical significance. Receiver operating Characteristic (ROC) curves were constructed and area under curve (AUC) was calculated for detection of the sensitivity and specificity of miRNAs for predicting HCC cases among HCV-infected patients. Correlation between the miRNAs expression levels and other variables was determined using Spearman’s correlation analysis.
The mean ages of the chronic hepatitis and HCC patients at diagnosis were 58.4 ± 8.5 years, 57.4 ±7.9 years. There was no significant difference among the mean ages of the HCV and HCC groups (P=0.256), (Tables 1-4) and (Figures 1-6).
Figure 1: Bar Chart presentation of the Child Pugh score across the two groups.
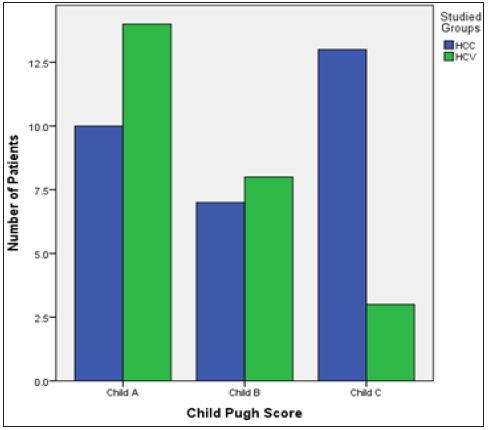
Table 1: demographic and laboratory variables presentation in the two groups.
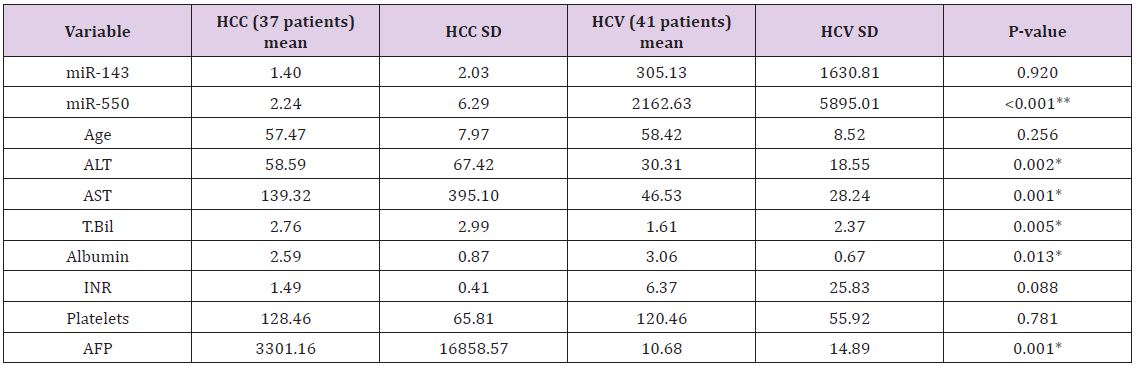
Figure 2: Micro RNA 143 level presented in box plot in the three groups.
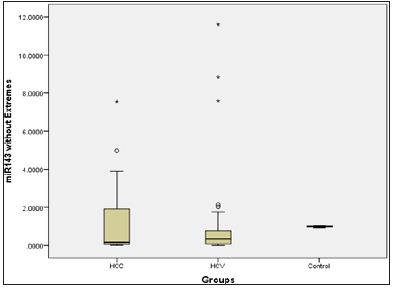
Figure 3: Micro RNA 550 level presented in a box plot in the three groups.
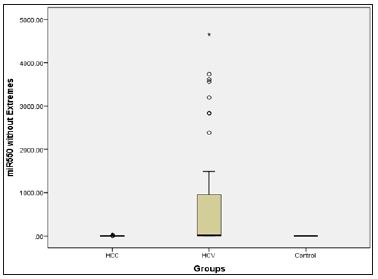
Figure 4: Box plot presentation of AFP in the HCV and HCC groups.
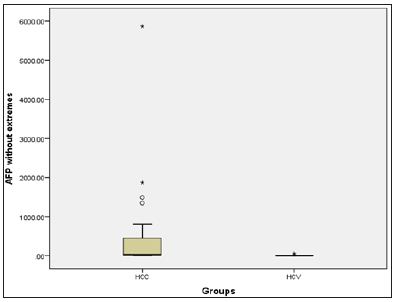
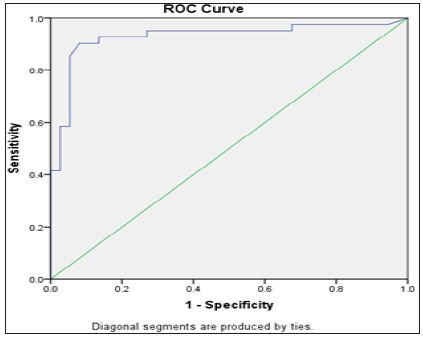
Figure 5: ROC curve of miRNA 550 in relation to the presence of HCC.
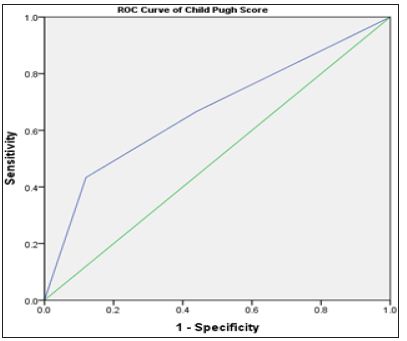
Biochemical Analysis: The laboratory data of the included patients showed that there was a significant difference between the HCV and HCC groups as regards the ALT, AST, Albumin, Total Bilirubin and AFP (Table 1). When using the Child Pugh score as a predictive value for the occurrence of HCC; it was shown that its AUC was 0.67, NPV=56.4 and PPV=81.2, with sensitivity 43.3% and specificity 88% at a cut-off score 10 i.e. Child C (Table 2).
Table 2: demographic and laboratory variables presentation in the two groups.
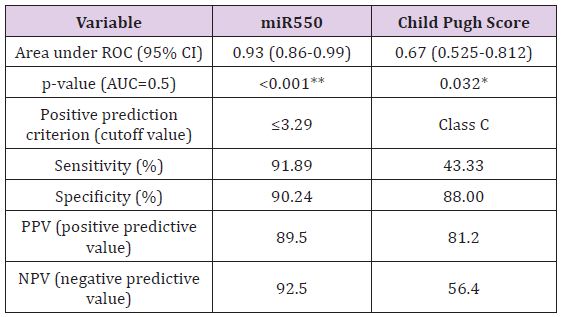
MiR-143 and miR-550a Expression in HCV and HCC and their use as Predictive Value for HCC Occurrence: Plasma miR- 143 should no significant difference between the two groups, while miR-550a was significantly decreased in the HCC group versus the HCV group. This reveals that the miR-550a could be down-regulated by HCC. The predictive ability of miR-550a was assessed using regression and the AUC was 0.93 with PPV=89.5 and NPV=92.5, the sensitivity of the test was 91.89% and its specificity was 90.24% at a cut off value 3.29. (Figure 5) & (Table 2) Correlation between the two microRNAs markers used and various laboratory parameters revealed the following; that miR- 143 was not correlated significantly with any laboratory parameter while miR-550a was correlated with AST, total bilirubin, albumin, INR, AFP and Child Pugh Score (Table 3). On the other hand, there was no relation between both markers and gender using the Mann Whitney test (Table 4).
Table 3: correlation between the microRNAs studied and various laboratory parameters.
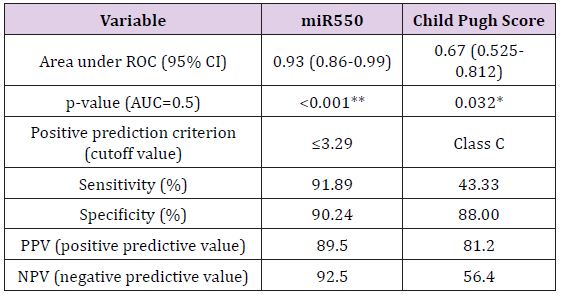
Table 4: Relation with Gender Mann-Whitney.

miR-143 is located in the chromosome 5 with a mature length of 22 [10]. A previous study has revealed that miR-143 exhibited down-regulation in many cancers such as colorectal cancer [11], pancreatic cancer [12] and lung cancer [13] in which miR-143 acts as a tumor suppressor inhibiting cell proliferation and inducing tumor cell apoptosis thus hindering the development of cancer [10]. miR-143 was exploited in many cancers e.g. lung cancer [14], gastric cancer [15], colorectal cancer [16], osteosarcoma [17], T-cell leukemia [18] and prostate cancer [19]. While some proposed the idea of a cancer suppression effect through mediators as IGF1 [11], matrix metalloprotease 13 [17] while others proposed the effect of miR-143 on ERK5 pathway increasing apoptosis [18,19] In the present study we found no correlation between miR-143 expression and occurrence of HCC in HCV infected patients, and this was in contrary to a recent study which showed that miR-143 was down-regulated in HCC patients [1].
First we have to mention that most studies that tackled this area concentrated mainly on the level of the micro RNA in the hepatocyte i.e. Histopathological and not the plasma level and this is an important differentiating point as serum level may be the higher with low cellular concentration and vice versa due to shedding from the cells. And to our knowledge all studies concerning miR-143 and miR-550a as markers for HCC used the hepatocyte level of miRNAs and not the serum form. This may because a great difference in results as the serum for could be affected by different factors like the amount of miRNA shedding from the cell or cellular destruction reflected on serum level, while factors like the actual cellular level may not be reflected. To our knowledge this is the first study to clear this point by measuring the serum not the cellar levels of miR- 143 and miR-550a.
Second, miR-143 results could be explained by the fact that miR- 143 was increased in patients that had recurrence or metastasis, as their study selected all stages of HCC, while our patients selection deferred in selecting patients with only non-metastatic HCC (although micro-metastasis could be present as apparent by the high AFP in some of the cases), so this could be a more appropriate marker for development of metastasis. In addition, it is important to note that while the difference in miR-143 was not significant between the HCV and HCC groups it was higher in the HCC group, which as revealed by the study mentioned above, the higher the miR-143 the better prognosis. On the other hand; this could also be explained by the hepatocyte destruction due to the HCC and hence higher level in the blood. Autophagy gene 2B (ATG2B) was found to be a possible target for miR-143, where miR-143 Overexpression is associated with down regulation of ATG2B [13]; thus decreasing the autophagy responsible for the cancer cell survival and inducing apoptosis [20].
The first study to investigate the role of miR-143 in HCC was done recently where the researchers exploited the different cellular pathways that could play important role in the HCC pathogenesis and found that GATA6 was the downstream target for miR-143, where its Overexpression will oppose the cancer suppressor effect of miR- 143 thus increasing the tumor invasion in rats [21]. We found that the expression level of plasma miR-550a was significantly lower in HCC than HCV patients. This increased expression of miR-550a could be explained by its protective role in inhibiting cancer in the HCV group. A recent study revealed that; when injecting a mouse harboring cancer breast cells with miR-550a-p3, their metastatic ability is reduced. This was explained by the effect of miR-550a-p3 in down-regulating the ERK1 and ERK2 [22] (two important effector kinases in the oncogenic Ras/ERK pathway [23]).
In a study done by Tian et al. [24] on liver tissue with HCC they found that; miR-550a is responsible for the migration and vascular invasion of HCC cells, they proposed that this could be accomplished through its direct and functional target CPEB4, where the expression of the later is closely correlated with HCC treatment outcome [24] In addition, we conduct ROC analyses to detect the potential application of miR-550a in the diagnoses of HCC. Our results showed that miR-550a might be a potential serum biomarker for HCC. Moreover, The ROC curve analysis of miR550a showed the good sensitivity and specificity in differentiating patients with HCC from those with HCV (Figure 5) & (Table 2). Another important finding is that serum miR-550a showed a statistically significant positive correlation with AST, total bilirubin, albumin, INR, AFP and Child Pugh Score (Table 3).
Finally, the Child Pugh score was a potential diagnostic marker for HCC (Table 2) & (Figure 6). This is in accordance with a lot of previous studies which showed that HCC is an important cause of sudden deterioration of liver function [25]. In conclusion, we suggest that plasma miR-550a might serve as a non-invasive diagnostic and prognostic serum marker of HCC, miR-550a was better than miR-143 in early diagnosis of HCC, although miR-143 needs more study on metastatic HCC for the evaluation of its effect on prognosis. Our study is limited by the small sample size and large-scale studies are in need for validation of our results.
Figure 6: ROC curve of Child Pugh score in relation to the presence of HCC.