Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Shovit Ranjan1 and Praveen Kumar Sharma*2
Received: June 22, 2018; Published: June 27, 2018
*Corresponding author: Praveen Kumar Sharma, Centre for Life Sciences, Central University of Jharkhand, Brambe, Ranchi, Jharkhand, India
DOI: 10.26717/BJSTR.2018.06.001314
Introduction: Brain-derived neurotrophic factor (BDNF) plays a key role in promoting the survival of neurons in the nervous system. Polymorphism in the gene region 196G>A and 270C>T of BDNF has been studied for susceptibility to Parkinson’s Disease (PD) but results from different studies are inconclusive.
Objective: To carry out a meta-analysis and trial-sequential analysis of the previous studies to assess the relationship between the BDNF 196G>A and 270C>T polymorphism and PD risk.
Methods:The databases were searched for the studies concerning BDNF 196G>A and 270C>T polymorphism and its association with PD risk. The pooled odds ratios (ORs) along with 95% confidence intervals (95% CIs) were calculated for all the genetic models, from the selected case-control studies, by meta-analysis. The required information size was calculated using α = 0.05 (two sided), β = 0.20 (power 80%) and a relative risk reduction of 20%, by using trial sequential analysis (TSA)
Results: Results of present meta-analysis identified an association between recessive AA Vs GG+AG genotype and PD in Asian population but no association between BDNF 196 G/A polymorphism and PD in European population. On the other hand, association between BDNF 270 C/T allele in overall studies was observed for T Vs C allelic contrast and dominant TT+TC Vs CC genotype.
Conclusion: Our meta-analysis demonstrate that the evidence for associations between BDNF polymorphisms (G196A and C270T) and PD risk for few allele and genotype combinations are present but is ethnicity dependent.
Keywords: BDNF; G196A; C270T; Parkinson’s Disease; Polymorphism; Meta-Analysis; Trial Sequential Analysis
Parkinson’s disease (PD) is the most common neurodegenerative disorder, clinically characterized by resting tremor, rigidity, gait abnormalities, postural imbalance and bradykinesia [1]. These underlying pathological events in PD result from the death of dopamine- generating cells in the region of the midbrain [2]. Although the etiology of PD is not fully known, the studies had already shown the role of both genetic factors [3,4] and environmental factors [5] in the pathogenesis of PD [6]. The brain-derived neurotrophic factor (BDNF) gene, encoding a nerve growth factor, and promoting the survival of dopaminergic neurons in the substantia nigra, is highly expressed in the nervous system [7,8]. Decreased BDNF mRNA expression and protein have been observed in the substantia nigra of PD patients [9,10] making BDNF, an important candidate gene for PD risk. Based on these observations, several molecular epidemiological studies have investigated the association of BDNF G196A (rs6265) and C270T (rs56164415) Single Nucleotide Polymorphisms (SNPs) with PD risk in different populations (Table 1). However, the findings from these studies for the probable association between these two SNPs on the susceptibility of PD remain inconsistent.
So, in an attempt to resolve these contradictory results, we performed this meta-analysis by collecting and sorting previously published case-control studies to make a more comprehensive and convincing evaluation of the overall and ethnicity specific PD risk associated with this polymorphism, as well as to evaluate this polymorphism as potential marker for screening of PD. Here we report the largest and comprehensive systematic review and meta-analysis to date, which uses an extensive search of observational studies to calculate the association of BDNF gene polymorphisms G196A and C270T for PD risk. Moreover, we also conducted Trial Sequential Analysis (TSA) of all the published case-control studies in the hope of providing more precise evidence.
Preferred Reporting Items for Systematic Reviews and Meta- Analyses (PRISMA) 2009 guidelines for systematic review and meta-analysis and the Cochrane Collaboration definition of both terms were followed for this work [11,12]. Literature search was carried out within PubMed (Medline), EMBASE and Science Direct database up to June, 2018, using the keywords- bdnf, gene, patient, polymorphism and Parkinson disease. Then, potentially relevant publications and studies were retrieved by examining their titles and abstracts and matching the eligible criteria, as done in the previous meta-analysis study [13].
To facilitate the proper interpretation of results and to minimize heterogeneity, all eligible studies had to fulfill the following inclusion criteria like evaluation of BDNF gene 196 G>A and 270 C>T with PD risk; use of case-control or cohort studies; recruitment of pathologically confirmed PD patients and healthy controls; and availability of genotypic frequency both in case and control. Moreover, when the case- control study was included by more than one article using the same case series, then we selected the study that included the largest number of individuals. The major reasons for exclusion of studies were overlapping data, case-only studies; review articles, family-based studies and animal studies.
For each meta-analysis, the methodological quality assessment and data extraction were independently abstracted in duplicate using a standard protocol. Data accuracy was ensured using data-collection form according to the inclusion and exclusion criteria listed above. In case of discrepancy on any item of the data collected from the retrieved studies, the problem would be fully discussed to reach a consensus. Data extracted from each studies included the name of first author, year of publication, ethnicity, number of cases and controls, types of study and genotyping methods and frequencies of the case and control.
The meta-analysis examined the overall association and ethnicity specific association of the A and T allele with the risk of PD relative to the G and C allele respectively, the contrast of homozygotes AA vs GG; TT vs CC, the contrast of heterozygotes AG vs GG; TC vs CC, the recessive model for the A allele: contrast AA vs (AG+GG); TT vs (TC+CC), and the dominant model for the A allele: contrast (AA+AG) vs GG; (TT+TC vs CC). All associations were indicated as odds ratios (ORs) with the corresponding 95% confidence interval (CI). A pooled OR was then estimated based on individual ORs.
Hardy-Weinberg equilibrium (HWE) was examined in the control subjects using a goodness of fit chi-square test for each study, Odds ratio (OR) with corresponding 95 % confidence intervals (CI) was used to evaluate the association between the BDNF 196 G>A gene polymorphism and BDNF 270 C>T with PD risk separately. Heterogeneity was assessed by Chi-square based Q-Test [14]. If heterogeneity existed, then random effects model was used to calculate the overall pooled OR value [15]; otherwise, the fixed effect model was use [16]. Moreover, I2 statistics was used to quantify inter study variability. It ranges between 0% and 100%, where a value of 0% indicates no observed heterogeneity, and larger values indicate an increasing degree of heterogeneity [17]. The HWE was examined in the control subjects using a goodness-of-fit chi-square test for each study. Begg’s funnel plots and Egger’s regression test were undertaken to evaluate the potential publication bias [18]. P value less than 0.05 was judged significant. Publication bias was assessed by visual inspection of funnel plots in which the standard error of log (OR) of each study was plotted against its log (OR). An asymmetric plot suggests a possible publication bias. Funnel plot asymmetry was also assessed by the Egger’s linear regression test. The significance of the intercept was determined by the t-test (p < 0.05 was considered representative of statistically significant publication bias [19]. All the data analysis was performed using a comprehensive meta-analysis (CMA) V2 software (Biostat, USA).
According to Cochrane Handbook for systematic reviews of interventions, meta-analyses and systematic reviews are considered the best available evidence if all eligible trials are included. However, the best available evidence might not always be equal to strong sufficient evidence. It is well known that meta-analysis may result in increased risk of random errors when series of sparse data are analyzed and in reduplicative significance testing when new trials are updated in cumulative meta-analysis. Therefore, keeping mind on the issues raised above, we applied the TSA to increase the robustness of current conclusions by minimizing the random errors [20-22]. The methods of using TSA were based on the ‘User manual for Trial Sequential Analysis (TSA)’. In the study, TSA was used to control the risk of random error by calculating the required information size and an adjusted threshold for statistical significance to make a robust conclusion [20-23].
The required information size was calculated with the assumption of a plausible relative risk of 20% with low risk bias, and the overall 5% risk for a type I error (α), 20% risk for a type II error (β) were adopted [24]. Based on required information size and risk for type I and type II errors, TSA monitoring boundaries were built. When the cumulative Z-curve crosses the TSA monitoring boundary before the required information size is reached, a sufficient level of evidence might have been reached and further trials are not necessary. Otherwise, evidence to reach a conclusion is insufficient and further trials are necessary [25]. The software Trial Sequential Analysis Viewer (version 0.9.5.5 Beta) was used for the study and 95% CIs was adjusted for sparse data or repetitive testing, described as the TSA-adjusted 95% CIs.
The literature review identified a total of 17 studies eligible for inclusion in our analysis as described in Flow Chart (Figure 1). Based on our preliminary search criteria, a total of 307 studies were identified in PubMed (Medline), EMBASE and Science Direct using the keywords- bdnf, gene, patient, polymorphism, Parkinson disease and their combination. After careful review, finally, 17 potential studies were included. According to our inclusion criteria, 8 studies have not been included for estimating OR and 95% CI because they didn’t report genotypic frequency of healthy controls [26-33]. Finally, 17 eligible studies involving 4336 cases and 4457 controls were enrolled in the pooled analyses. The populations came from 12 different countries, including China, Colombia, Finland, Greece, Italy, Japan, Poland, Serbia, Spain, Sweden, Taiwan and USA. Detailed characteristics of all eligible studies included in meta-analysis are reported in (Table 1). One overall study was conducted on BDNF 196GA polymorphism and 2 ethnicity specific studies were conducted, that includes 8 studies on Asian population [34-41], 6 studies on European populations [42-47]. Moreover, one overall study was also conducted on BDNF 270CT polymorphism. Ethnicity specific studies were not possible in this polymorphism due to less number of works. Table 2 and (Table 3) reports genotypic distribution of G196A and C270T polymorphism of BDNF gene from each study. All studies observed HWE.
Overall, the meta-analysis results based on different genetic models (Allelic, Homozygote, Heterozygote, Dominant and Recessive) revealed no association between BDNF 196 G/A allele in overall studies except for recessive AA vs AG+GG genotype. However, ethnicity specific meta-analysis identified an association between recessive AA vs AG+GG genotype and PD in Asian studies but no association identified between BDNF 196 G/A polymorphism and PD in European studies. The pooled ORs of overall study analysis revealed that BDNF G>A gene polymorphism is associated with PD risk in recessive (AA vs AG+GG: p = 0.046; OR = 1.265, 95% CI = 1.005 to 1.593) genetic models but not associated with PD risk in allelic (A vs G: p = 0.189; OR = 1.091, 95% CI = 0.958 to 1.243) genetic models; homozygous (AA vs GG: p = 0.147; OR = 1.216, 95% CI = 0.934 to 1.585) genetic models; heterozygous (AG vs GG: p = 0.826; OR = 0.977, 95% CI = 0.795 to 1.201) genetic models; and dominant (AA+AG vs GG: p = 0.621; OR = 1.047, 95% CI = 0.872 to 1.259) genetic models (Figure 2). All ORs were pooled through a random effect models (Table 4).
Figure 2: orest-Plot of a meta-analysis of the association between BDNF gene 196 G>A polymorphism (A vs. G; AA vs. GG; AG vs. GG; AA+AG vs. GG; AA vs. AG+GG) and overall PD risk.
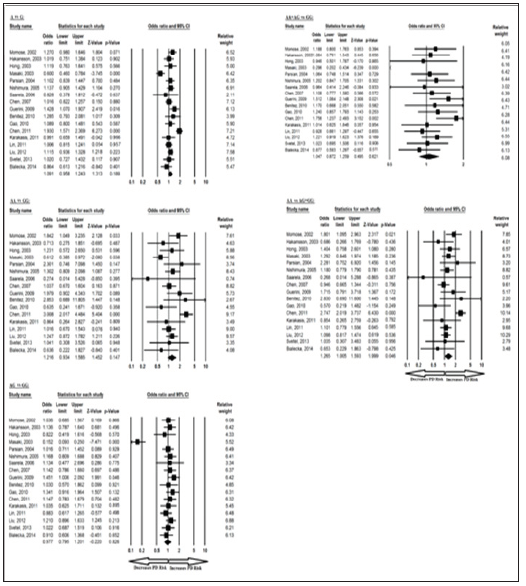
Similarly, the pooled ORs of Asian study analysis revealed that BDNF G>A gene polymorphism is associated with PD risk in recessive (AA vs AG+GG: p = 0.030; OR = 1.362, 95% CI = 1.030 to 1.802) genetic models but not associated with PD risk in allelic (A vs G: p = 0.377; OR = 1.104, 95% CI = 0.886 to 1.375) genetic models; homozygous (AA vs GG: p = 0.160; OR = 1.280, 95% CI = 0.907 to 1.807) genetic models; heterozygous (AG vs GG: p = 0.398; OR = 0.839, 95% CI = 0.558 to 1.261) genetic models; and dominant (AA+AG vs GG: p = 0.952; OR = 0.989, 95% CI = 0.691 to 1.415) genetic models (Figure 3). All ORs were pooled through a random effect models (Table 5). Moreover, the pooled ORs of European study analysis revealed that BDNF G>A gene polymorphism is not associated with PD risk in allelic (A vs G: p = 0.388; OR = 1.066, 95% CI = 0.922 to 1.232) genetic models; homozygous (AA vs GG: p = 0.928; OR = 1.021, 95% CI = 0.656 to 1.588) genetic models; heterozygous (AG vs GG: p = 0.217; OR = 1.117, 95% CI = 0.937 to 1.331) genetic models; dominant (AA+AG vs GG: p = 0.274; OR = 1.099, 95% CI = 0.928 to 1.301) genetic models; and recessive (AA vs AG+GG: p = 0.900; OR = 0.972, 95% CI = 0.628 to 1.507) genetic models (Figure 4). All ORs were pooled through a fixed effect models (Table 6).
Figure 3: Forest-Plot of a meta-analysis of the association between BDNF gene 196 G>A polymorphism (A vs. G; AA vs. GG; AG vs. GG; AA+AG vs. GG; AA vs. AG+GG) and Asian PD risk.
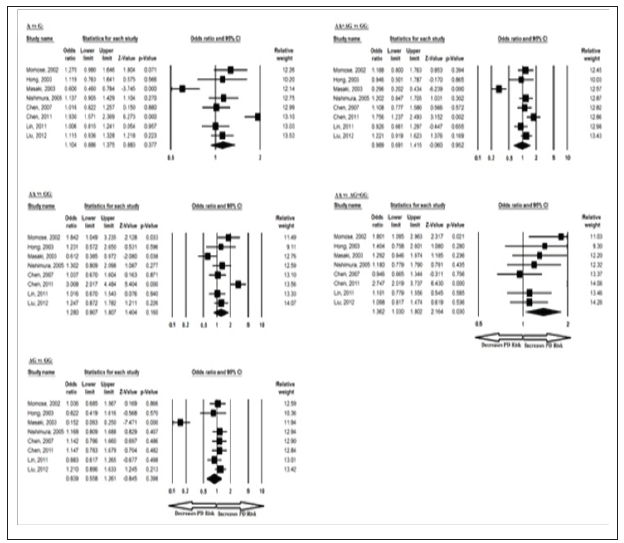
Figure 4: Forest-Plot of a meta-analysis of the association between BDNF gene 196 G>A polymorphism (A vs. G; AA vs. GG; AG vs. GG; AA+AG vs. GG; AA vs. AG+GG) and European PD risk.

The meta-analysis results based on different genetic models revealed association between BDNF 270 C/T allele in overall studies for T vs C allelic contrast and dominant TT+TC vs CC genotype. Ethnicity specific studies were not possible in this case because of less number of studies. The pooled ORs of overall study analysis revealed that BDNF C>T gene polymorphism is associated with PD risk in allelic (T vs C: p = 0.017; OR = 1.373, 95% CI = 1.059 to 1.778) genetic models and dominant (TT+TC vs CC: p = 0.026; OR = 1.368, 95% CI = 1.038 to 1.803) genetic models but not associated with PD risk in homozygous (TT vs CC: p = 0.097; OR = 3.925, 95% CI = 0.781 to 19.732) genetic models; heterozygous (TC vs CC: p = 0.051; OR = 1.321, 95% CI = 0.999 to 1.746) genetic models; and recessive (TT vs TC+CC: p = 0.102; OR = 3.837, 95% CI = 0.764 to 19.257) genetic models (Figure 5). All ORs were pooled through a fixed effect models (Table 7).
Figure 5: Forest-Plot of a meta-analysis of the association between BDNF gene 270 C>T polymorphism (T vs. C; TT vs. CC; TC vs. CC; TT+TC vs. CC; TT vs. TC+CC) and overall PD risk.
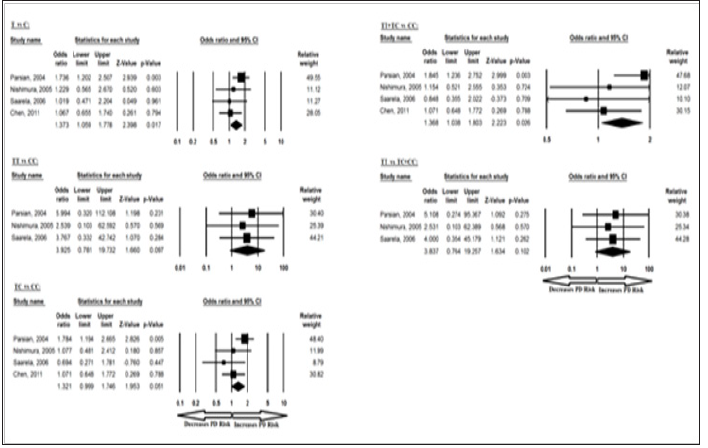
Table 7: Statistics to test publication bias and heterogeneity in meta-analysis (rs56164415-Overall).
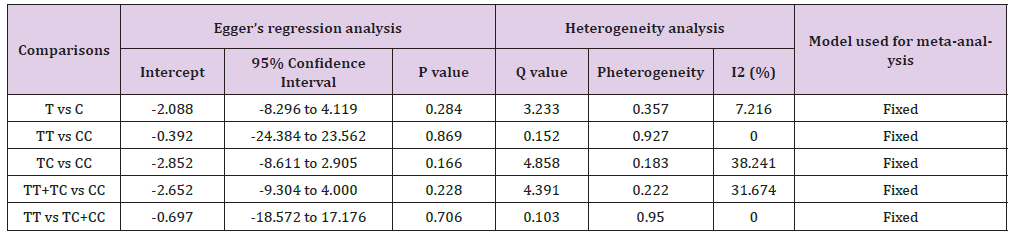
No between-study heterogeneity was found in analyses of the BDNF 196 G/A and 270 C/T polymorphisms in the overall, Asian or European study populations. Begg’s Funnel Plot and Egger’s Test were performed to evaluate the publication bias among the included studies for this meta-analysis. The shape of funnel plots did not reveal any evidence of obvious symmetry in all comparisons and the Egger’s regression test was used to provide statistical evidence of funnel plot. The results of Egger’s regression analysis did not show any evidence of publication bias in all genetic models (Egger’s regression test p values > 0.05; (Tables 4-7).
Sensitivity analysis was conducted to verify the robustness of our results. It is also used to ascertain whether modification of the inclusion criteria of the meta-analysis affected the final results. The effect of each study included in this meta-analysis assessed by sensitivity analysis of each individual study on the pooled OR by eliminating each single case-control study was done for each BDNF polymorphism [rs6265(G>A), rs56164415(C>T)] to evaluate the influence. Outcomes of sensitivity analysis revealed that no individual genetic model influenced the pooled ORs significantly in all the BDNF variants, which suggest the credibility and stability of this meta-analysis.
Seventeen trials (10768 subjects) were used to investigate the association of rs6265 and rs56164415 gene polymorphisms with PD risk. Using the data of recessive model for rs6265 (including 17 trials with 8793 subjects) as an example, the TSA was performed and found that the required information size (RIS) is 11795 subjects to demonstrate the issue. The cumulative Z-curve crosses the TSA monitoring boundary before reaching RIS, indicating that the cumulative evidence is sufficient and further trials are not necessary (Figure 6). However, the cumulative Z-curve does not crossed with TSA monitoring boundary when we performed the analysis using the data of dominant model, confirming that cumulative evidence is insufficient and further relevant trials are necessary (Figure 7). Similarly, when we performed the sub-analysis based on the ethnicity (Asian and European) for all models, the cumulative Z-curve does not crossed with TSA monitoring boundary except for recessive model of Asian, confirming that cumulative evidence is insufficient and further relevant trials are necessary (figures were not shown). Moreover, for rs56164415, we chose the data of four models to perform TSA. The cumulative Z-curve have not crossed with TSA monitoring boundaries before the required information size is reached, indicating that cumulative evidence is insufficient and further trials are necessary (figures were not shown).
Figure 6: Trial sequential analysis of 17 studies (using the data of recessive model) to demonstrate the relevance of rs6265 gene polymorphisms with PD susceptibility. The required information size was calculated using α = 0.05 (two sided), β = 0.20 (power 80%) and a relative risk reduction of 20%. The solid blue line represents the cumulative Z-curve.
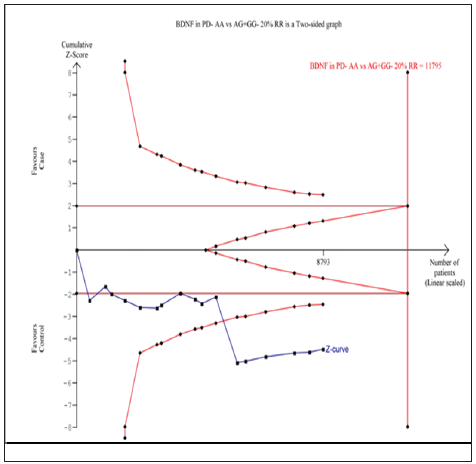
Figure 7: Trial sequential analysis of 17 studies (using the data of dominant model) to demonstrate the relevance of rs6265 gene polymorphisms with PD susceptibility. The required information size was calculated using α = 0.05 (two sided), β = 0.20 (power 80%) and a relative risk reduction of 20%. The solid blue line represents the cumulative Z-curve.

Even though the multi factorial nature of PD is well known, genetic factors are considered to be strong causes of the disease and, in view of that, various genes have been studied for the same. One such gene is BDNF [6], a member of the neurotrophin family of growth factors, which enhances the survival of dopaminergic neurons in the substantia nigra, and whose expression gets decreased in this region in PD patients [48,49]. Mutation in BDNF gene may affect the normal function of protein and increase the PD risk. Common genetic polymorphisms in the BDNF gene may alter protein function and play a major role in PD. This includes BDNF 196 G/A polymorphism, which generates Valine to Methionine amino acid substitution in the amino terminus of BDNF, resulting in abnormal intracellular distribution and decreased BDNF secretion [50]. In addition to this, BDNF 270 C/T polymorphism has also been studied to have an effect on BDNF function [51]. In the recent years, interest in the genetic susceptibility to PD has led to a growing attention to the study of gene polymorphisms involved in it. Several case-control studies have supported an important role for genetics in determining the risk for PD, and association studies are appropriate for searching susceptibility genes involved in PD [52].
Till date, series of epidemiological studies have been performed to explore the role of BDNF gene 196 G>A and 270 C>T polymorphism on PD susceptibility in worldwide population, but the results remain controversial and inconclusive. Some studies are limited by their sample size and subsequently suffer from too low power to detect effects that may exist. Meta-analysis is a powerful tool for summarizing the results from different studies and gives more reliable results than a single case-control study, where individual sample sizes are small and inadequate statistical power [53]. Combining data from many studies has the advantage of reducing random error [54]. So, in order to explore these contradictory findings, improve the statistical power and determine the effect size of BDNF gene 196 G>A and 270 C>T polymorphism, the authors conducted a meta- analysis with seventeen eligible studies to provide the more comprehensive and reliable association between BDNF gene 196 G>A and 270 C>T polymorphism and overall PD risk for worldwide, Asian or European population by combining data from all the available case-control studies on the topic published till now.
Summarizing the clinical data available by 17 studies, results of the present meta-analysis showed that BDNF gene 196 G>A and 270 C>T polymorphism may be significantly associated with increased PD risk in overall population. Subjects with recessive AA vs AG+GG genetic model for BDNF 196 G>A polymorphism in overall and Asian populations had 1.26- and 1.36-fold increased risk of developing PD as compared with the wild genotype, respectively. But, allelic, homozygous, heterozygous and dominant models for BDNF 196 G>A polymorphism in both overall and Asian population have not found to be associated with the risk of PD. Moreover, allelic, homozygous, heterozygous, dominant and recessive genetic models for BDNF 196 G>A polymorphism in European populations has also found to be not associated with the risk of PD. On the other hand,subjects with C allele and dominant TT+TC vs CC genetic model for BDNF 270 C>T polymorphism in overall populations had 1.37- and 1.36-fold increased risk of developing PD as compared with the wild T allele and genotype, respectively. But, homozygous, heterozygous and recessive models for BDNF 270C>T polymorphism in overall population have not found to be associated with the risk of PD.
Based upon the above results and importance of BDNF’s role in the pathogenesis of PD, it is biologically plausible that BDNF gene 196G>A and 270C>T polymorphism may modulate the risk of PD and could be a genetic factor for inter-individual differences in susceptibility to PD. Since, genetic susceptibility to PD is polygenic type [55], therefore, single genetic variant might be inadequate to predict the risk of this fatal disease. Some limitations should be addressed which might affect the result, i.e., first, inter-study heterogeneity found in this meta-analysis due to many factors like different regional lifestyle among populations from different parts of world; recruitment of control group, as the controls were not uniformly defined- some studies used a healthy population as the reference group where as other selected hospital patients without PD as the reference group. Second, the present meta-analysis was based mainly on unadjusted effect estimates and Confidence Intervals (CIs). Third, the gene-gene and gene- environment interactions were not addressed. Regardless of the above stated limitations, there are some advantages associated with this meta-analysis. First this meta-analysis included more number of studies compared to previously published pooled analysis with increased statistical power and resulted statistically significant and robust conclusion. Second, Funnel plots and Egger’s tests for this meta-analysis did not detect any potential publication bias. Also, the supplementary sensitivity analysis supported that the results of the present metaanalysis are highly stable and reliable.
This comprehensive systematic review and meta-analysis revises the previous incomplete data and indicates that BDNF G196A and C270T polymorphism would be a risk factor for PD susceptibility. The importance of this polymorphism as a predictor of the risk of PD is very high and the screening utility of this genetic variant in symptomatic individuals may be warranted. So, future well designed large scale and multi- ethnicity studies with homogeneous PD patients and well-matched controls might be necessary to investigate the association between BDNF gene SNPs and risk of PD.
This work was partially supported by a grant from the Department of Biotechnology, Government of India BUILDER project (BT/ PR-9028/INF/22/193/2013) to Centre for Life Sciences, and UGC Central University Fellowship to S.R.


