Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Marina Tusup1,2, Lars E French1,2, Emmanuella Guenova1,2, Thomas Kundig1,2, and Steve Pascolo1,2*
Received: July 22, 2018; Published: July 27, 2018
*Corresponding author: Steve Pascolo, University Hospital of Zürich, Department of Dermatology, Gloriastrasse 31, 8091 Zürich, Switzerland
DOI: 10.26717/BJSTR.2018.07.001487
Background: in vitro-transcribed messenger RNA (ivt mRNA) is a safe genetic vector that can be used for vaccination and gene therapy. We investigated the impact of the 3’ untranslated region (UTR) and of modified nucleotides on the functionality of ivt mRNA in vitro as well as in vivo. We confirmed that a 3’ UTR consisting of a tandem repeat of beta-globin 3’ UTR enhances the expression of ivt mRNA in non-transformed cells in vitro and in liver in vivo. In addition, we compared ivt mRNA made with the four canonical bases (“ACGU”), with uridine replaced by N1-methyl pseudouridine (“ACGPseudo”) or with both uridine and cytidine replaced by N1-methyl pseudouridine and 5-methylcytidine, respectively (“A5mCGPseudo”). We report that the “ACGU” and “ACGPseudo” ivt mRNA have superior functionality in tumour cells. However, in immune cells and in vivo, the “ACGPseudo” composition allows better expression of the synthetic mRNA. The additional substitution of cytidine with 5-methylcytidine in “A5mCGPseudo” ivt mRNA is deleterious for expression. The “ACGU” ivt mRNA induces cytokines in immune cells, while both “ACGPseudo” and “A5mCGPseudo” do not. We conclude that an ivt mRNA containing A, C, G and N1-methyl pseudouridine residues and having a 3’ tandem repeat of the beta-globin UTR is the optimal design of this vector for gene therapy.
Keywords:in vitro-transcribed mRNA, Pseudouridine, 5-methylcytidine, Globin UTR
The utilization of in vitro-transcribed messenger RNA (ivt mRNA) for vaccination (induction of an immune response against the encoded protein) and gene therapy (expression of a protein without triggering innate and adaptive immune response) is now intensively developed [1,2]. Since 2006, the production of ivt mRNA at a large scale and under good manufacturing practice (GMP) is well established and has made possible the clinical testing of ivt mRNA in patients with melanomas or carcinomas [3-6]. Features that affect the efficacy of ivt mRNA include the capacity of the 5’ untranslated region (UTR) to recruit initiation factors and ribosome subunits, of the coding region to be efficiently translated and of the 3’ UTR to stabilize the mRNA. In addition to its primary function of serving as a matrix for protein production, ivt mRNA is a danger signal [7]. It can trigger different types of immune responses, depending, for example, on the size of the particles with which it is associated [8].
Although the induction of innate immunity by ivt mRNA can be a good adjuvant in the context of vaccines, it can also limit expression (type I interferon blunts the translation machinery) and prevent the utilization of this format for gene therapy. Substituting canonical unmodified nucleotides such as uridine and cytidine with modified nucleotides such as pseudouridine () and 5-methylcytidine (5mC), respectively, was shown to reduce the immunogenicity of synthetic RNA [9]. It was also reported that those two substitutions can increase the translation of mRNA in immune (e.g., dendritic cells) and cancer cell lines (HEK293T) [10,11]. Synthetic mRNA containing N1-methyl pseudouridine (1) was found to outperform in a broad range of cells (the tumour cell lines A549 and HeLa, human fibroblast BJ cells and human primary keratinocytes) [12].
Having implemented an academic ivt mRNA production and formulation platform in Zurich (http://www.cancer.uzh.ch/en/Research/mRNA-Platform.html) we aimed to further understand and optimize the functionalities of this gene therapy format. Previously published work used ivt mRNA that was either co-transcriptionally capped with the ARCA analogue or post-transcriptionally capped using vaccinia capping enzymes. We here use mRNA co-transcriptionally capped with a newly available superlative CAP1 analogue: a dinucleotide called CleanCapTM AG that has the formula m7G(5’)ppp(5’)(2’OMeA)pG (TriLink). Concerning the 5’ UTR, we recently found that a sequence corresponding to an eIF4G aptamer was capable of enhancing translation of the ivt mRNA [13]. We here investigated the 3’ UTR and the impact of unmodified versus modified residues on the functionality of CleanCapTM ivt mRNA.
PCR DNA fragments with a modified T7 promoter sequence (TAA TAC GAC TCA CTA TAA GG) and an eiF4G aptamer (ACT CAC TAT TTG TTT TCG CGC CCA GTT GCA AAA AGT GTC G) followed by a codon- optimized firefly luciferase-coding gene (a synthetic gene purchased from Blue Heron Bio) or ZsGreen gene (Clontech) and terminated by either the sequence TAA or 5’ GAGAGCTCGCTTTCTTGCTG 3’ or a tandem repeat of the beta-globin 3’ UTR 14 were used as matrices for mRNA production with a HiScribe™ T7 mRNA Kit (New England Biolabs). The nucleotide mixture consisted of 8 mM CleanCapTM (a newly available CAP1 analogue), ATP, GTP, either CTP or 5mCTP and either UTP or N1-methyl pseudouridine (1) triphosphate (all from TriLink) at a final volume of 20 microlitres.
After incubation for 3 hours at 37°C, 80 microlitres of a solution containing 10 microlitres of 10x buffer, 3 microlitres of poly (A) polymerase, 1 microlitre of DNase and 1 microlitre of RNasin (all from New England Biolabs) were added. The reaction was incubated for 3 hours before the addition of 50 microlitres of 8M LiCl, cooling at -20°C for 30 minutes and centrifugation at 15000 rpm for 15 minutes. Supernatants were removed, and 200 microlitres of 75% ethanol was added. After a 10 minute centrifugation at 15000 rpm, the supernatants were discarded, and the RNA pellets were dried in a “Speed-Vac”. RNAs were then resuspended in water, and the concentration measured using a Nanodrop was adjusted to 1 microgram per microlitre. Quality and integrity of ivt mRNAs were checked using agarose gel electrophoresis. The RNAs were stored at -20°C.
Human embryonic kidney (HEK) cells and CT26 mouse colon carcinoma cells were maintained in RPMI medium (Thermo Fisher Scientific) containing 10% foetal calf serum (FCS) and 0.2% antimicrobial reagent Normocin (InvivoGen). Human peripheral mononuclear blood cells (PBMCs) and mouse splenocytes were obtained from peripheral blood from healthy donors and wild type C57Bl/6 or BALB/c mice, respectively, using centrifugation with Ficoll. For Luciferase experiments, transfection of tumour cells (CT26 or HEK) was performed with 100 000 tumour cells per well in 100 microlitres of RPMI medium supplemented with 10% FCS and 0.2% antimicrobial reagent Normocin (Invivogen) by adding a mixture of 20 ng of mRNA in 0.5 microlitres of Opti- MEM (Thermo Fisher Scientific) and 40 ng of the lipofectamine transfection reagent MessengerMAX (Thermo Fisher Scientific) in 0.5 microlitres of Opti-MEM to each well.
Transfection of non-transformed cells (PBMCs or splenocytes) was performed with 1 million cells per well in 100 microlitres of RPMI medium supplemented with 10% FCS and 0.2% antimicrobial reagent Normocin (Invivogen) by adding a mixture of 200 ng of mRNA in 5 microlitres of Opti-MEM and 400 ng of MessengerMAX in 5 microlitres of Opti-MEM to each well. Luciferase activity was recorded one day after transfection by adding 25 microlitres of Bright-Glo luciferase assay solution (Promega) and measuring activity using GloMax luminometer equipment (Promega). For ZsGreen experiments, transfection of tumour cells (CT26 or HEK) was performed with 100 000 tumour cells per well in 200 microlitres of RPMI medium supplemented with 10% FCS and 0.2% antimicrobial reagent Normocin (Invivogen) by adding a mixture of 100 ng of mRNA in 2.5 microlitres of Opti-MEM (Thermo Fisher Scientific) and 200 ng of the lipofectamine transfection reagent MessengerMAX (Thermo Fisher Scientific) in 2.5 microlitres of Opti-MEM to each well. Fluorescence (Excitation: 485nm; Emission: 528nm) was recorded in real time at 37°C using Cytation™ 3 Cell Imaging Multi-Mode Reader (BioTek). Immunostimulation experiments were performed by forming protamine-RNA nanoparticles as previously described 15 (mixing RNA at 0.5 mg/ ml in water with protamine at 0.5 mg/ml in water) and incubating them with PBMCs for 20 hours at a final RNA concentration of 5 microgram/ml. Then, the interferon-alpha and tumour necrosis factor (TNF)-alpha contents in the cell culture supernatants were recorded using ELISAs (from Mabtech and eBioscience, respectively).
Our study “Anti-cancer therapies based on RNA” was approved by the Veterinary Office of the University of Zurich (Kanton Zürich, Health Direction, Veterinary Office, Zollstrasse 20; 8090 Zurich; license number ZH215/17). The Veterinary Office of the University of Zurich has a research ethics review committee that granted approval. All efforts were made to minimize suffering. Animals were purchased from Envigo (Netherlands). Mice of 4 to 8 weeks of age were injected intra-venously with 1 microgram of mRNA formulated with a TransIT-mRNA transfection kit following the manufacturer’s suggestions (Mirus). Briefly, per mouse, 1 microgram of mRNA was diluted in 38 microlitres of Opti-MEM (Gibco) before the addition of 0.72 microlitres of “mRNA Boost Reagent”. After mixing, 1.12 microlitres of TransIT reagent was added. The solution was thoroughly homogenated and injected immediately. Five hours later, bioluminescence in vivo imaging was performed on an IVIS Lumina instrument (PerkinElmer). Immediately before the measurements, D-luciferin (Synchem) dissolved in PBS (15 mg/ml stock) and sterile filtered was injected (150 μg/g intraperitoneally). Emitted photons from live animals were quantified 10-20 minutes post luciferin injections, with an exposure time of 1 min. Regions of interest (ROI) were quantified for average radiance (photons s−1 cm−2 sr−1) (IVIS Living Image 3.2).
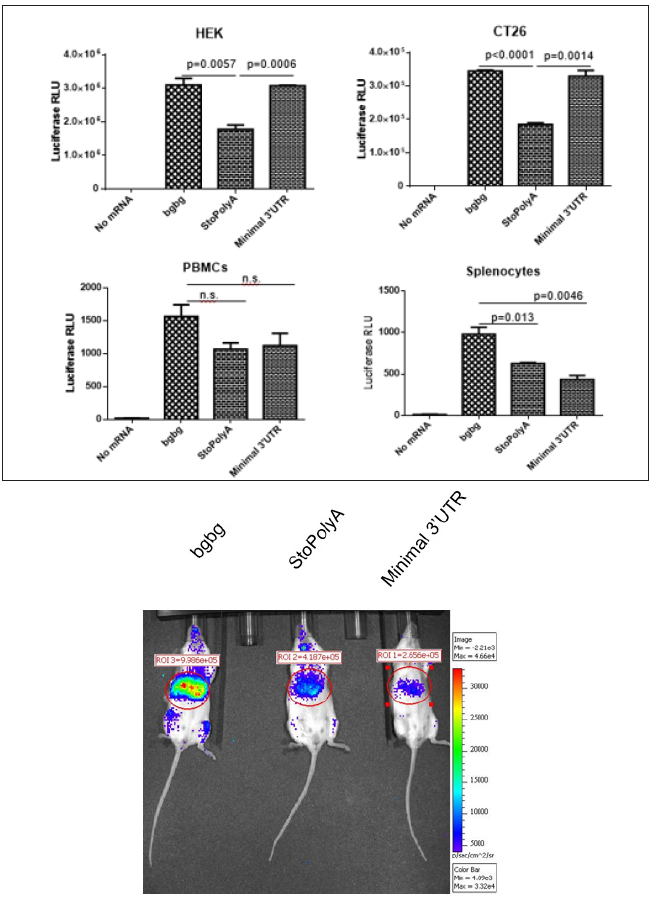
Using a transcription reaction containing ATP, GTP, 5mC and 1 triphosphate, we generated luciferase-coding mRNAs that had a unique optimized 5’ UTR (a “CleanCapTM” cap structure and an eIF4G aptamer sequence [13] and a codon-optimized coding sequence terminated at the 3’ end with a stop codon, TAA, immediately followed by a poly (A) sequence (“StoPolyA”), a stop codon followed by the minimal sequence “5’ GAGAGCTCGCTTTCTTGCTG 3’” and then a poly (A) tail (“Minimal 3’ UTR”) or a stop codon followed by a tandem repeat of the beta-globin 3’ UTR and a poly (A) tail (“bgbg”), as described previously [14]. Those mRNAs were transfected into cells, and the luciferase activity was recorded one day later.
As presented in Figure 1A, all three types of ivt mRNAs could be translated in cells. In tumor cells such as human embryonic kidney (HEK) cells and undifferentiated mouse colon carcinoma (CT26 cells) cells, the presence of a sequence between the stop codon and the poly (A) tail, whether a short or double repeat of beta-globin UTR, performed best. However, in immune cells, such as human peripheral blood mononuclear cells (PBMCs) or mouse splenocytes, the mRNA constructs with a double beta-globin 3’ UTR allowed a higher expression of luciferase. The advantage provided by the “bgbg” 3’ UTR was even more apparent in vivo in mice administered intravenously with mRNA formulations allowing expression primarily in the liver and in lymphoid organs (Figure 1B). The luciferase signal seen at 5 hours post intravenous injection, which corresponds to the reported peak of expression of this method [16], was stronger when using the “bgbg” ivt mRNA than when using the other ivt mRNAs. At 24 hours post injection, a luciferase signal could no longer be detected (data not shown). Thus, it can be concluded that an optimized CleanCapTM ivt mRNA can be obtained by adding to its 3’ UTR a tandem repeat of the betaglobin 3’ UTR, as previously discussed [14].
Figure 1: Effects of different 3’ UTRs on the functionality of ivt mRNA.
Synthetic mRNA with a tandem repeat of the beta-globin 3’ UTR (“bgbg”), no nucleotide (“StoPolyA) or a short sequence (“Minimal 3’ UTR”) between the stop codon and the poly (A) tail were (A) transfected using MessengerMAX in tumour cells (human HEK and mouse CT26 cells) or immune cells (human PBMCs or mouse splenocytes) or (B) injected using a formulation that allows expression of the ivt mRNA primarily in the liver. The results in A present the average and standard deviation (from triplicates) of the luciferase activity at 22 hours post transfection (p values using unpaired t-test are indicated; n.s. means not significant), and the results in B present luciferase expression in vivo 5 hours after intra-venous injection (the numbers indicate radiance in the region of interest). Red indicates high while blue indicates low radiance.
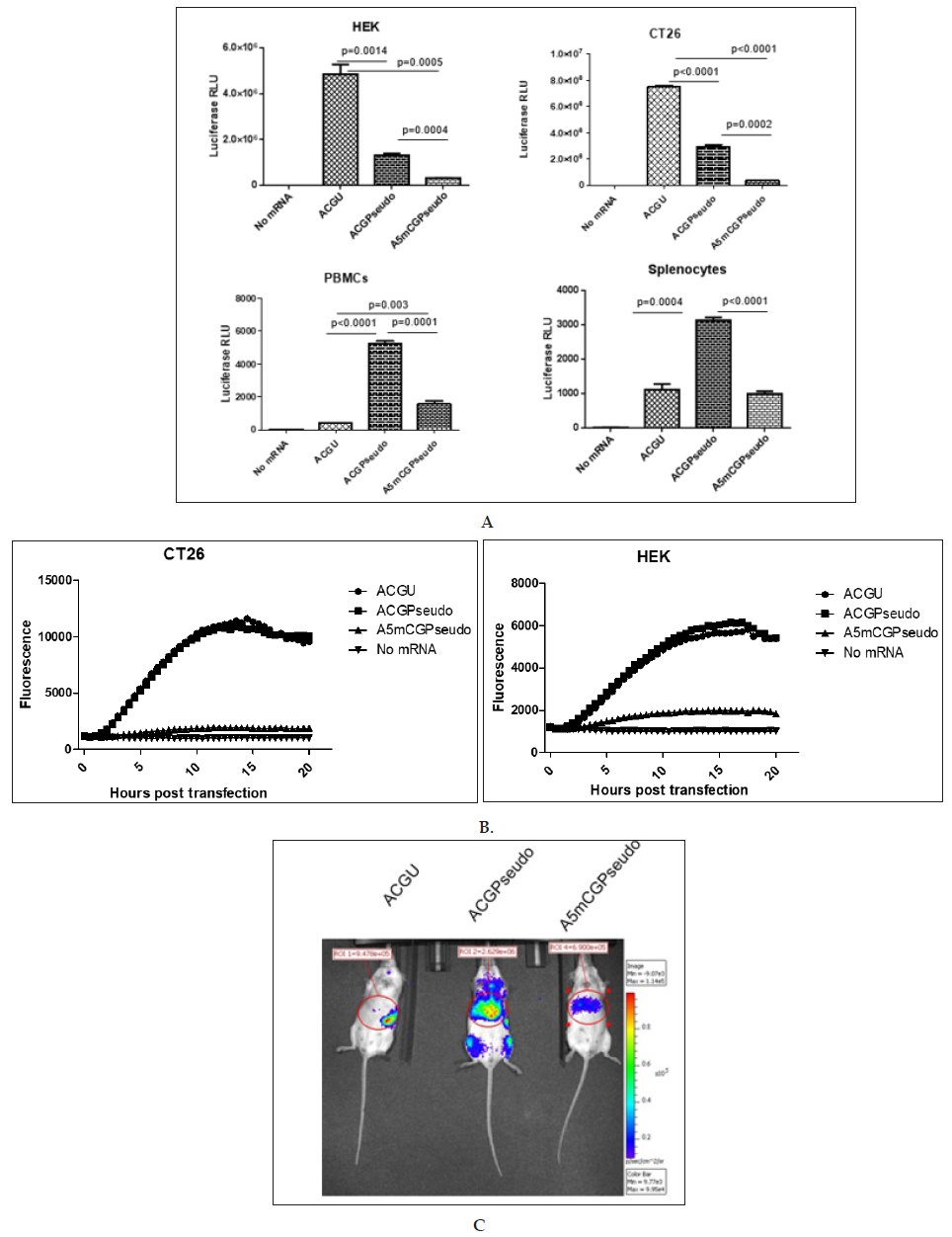
Using a PCR matrix with a 5’ eIF4G aptamer sequence [13], a codon-optimized coding sequence and a tandem repeat of the betaglobin 3’ UTR, we generated ivt mRNAs containing either A, C, G and U residues (“ACGU”); A, C, G, and 1 residues (“ACGPseudo”) or A, 5mC, G, and 1 residues (“A5mCGPseudo”). All RNAs had a CleanCapTM cap structure and were polyadenylated using a poly (A) polymerase. They were transfected into cells, and the luciferase activity was recorded one day later. As presented in (Figure 2A), the ACGU mRNA performed best in tumour cells, while the ACGPseudo mRNA performed best in immune cells. Using real-time fluorescence measurement ACGU and ACGPseudo mRNAs showed similar functionality (Figure 2B). This pattern reveals that
a) the functionality of CleanCapTM ivt mRNA might be reduced by the introduction of 1 and 5mC (differing from previous reports that indicate that those substitutions in posttranscriptionally capped ivt mRNA enhance functionality in tumour cell lines [10-12]) and
b) the triggering of immune receptors in immune cells by unmodified ivt RNA induces a state of reduced translation (the “interferon” effect) that abrogates expression from the transfected ivt mRNA more strongly than do modified nucleotides. This effect was visible in vivo at the peak of expression (ca. 5 hours [16]): after injection of mRNA formulated to be delivered primarily to the liver, the “ACGU” ivt mRNA was not expressed in the liver but was slightly expressed in the spleen, probably by immune cells such as CD169+ macrophages resistant to the interferon effect [17], while “ACGPseudo” and “A5mCGPseudo” ivt mRNAs were expressed primarily in the liver, with the former being approximately four-fold higher than the latter (Figure 2C). Imaging at 24 hours post injection did not allow detection of any signal (data not shown).
Figure 2: Effects of modified nucleotides on the functionality of ivt mRNA.
Synthetic mRNA with a 5’ UTR consisting of a CleanCap followed by an eIF4G aptamer, a codon-optimized coding region and a tandem repeat of the beta-globin 3’ UTR (“bgbg”) followed by a poly (A) tail was produced in the presence of the 4 canonical nucleotides (“ACGU”), with a full substitution of uridine by N1-methyl pseudouridine (“ACGPseudo”) or with a full substitution of both uridine and cytidine by N1-methyl pseudouridine and 5-methylcytidine, respectively (“A5mCGPseudo”). The ivt mRNAs were (A and B) transfected using MessengerMAX in tumour cells (human HEK and mouse CT26 cells) or immune cells (human PBMCs or mouse splenocytes) or (C) injected using a formulation that allows expression of the ivt mRNA primarily in the liver. The results in A present the average and standard deviation (from triplicates) of the luciferase activity at 20 hours post transfection of ivt mRNA coding luciferase (p values using unpaired t-test are indicated; n.s. means not significant), the results in B present the fluorescence that develops in the cells transfected with ivt mRNA coding ZsGreen over time and the results in C present luciferase expression in vivo 5 hours after intra-venous injection of ivt mRNA coding luciferase (the numbers indicate radiance in the region of interest). Red indicates high while blue indicates low radiance.
The capacity of “ACGU”, “ACGPseudo” and “A5mCGPseudo” ivt mRNAs to trigger innate immunity was investigated using the strongly immunostimulating protamine-RNA formulation [15]. While “ACGU” ivt mRNA induced a strong production of both interferon-alpha and TNF-alpha, neither the “ACGPseudo” nor the “A5mCGPseudo” ivt mRNAs triggered immunity (no significant differences between “No mRNA” or “Protamine” when compared to “ACGPseudo” or “A5mCGPseudo” mRNA for both interferon-alpha and TNF-alpha) (Figure 3). Thus, using production and purifications methods described here, the substitution of only U residues by 1 residues was sufficient to fully abrogate the danger signal feature of ivt mRNA.
We report here that a superlative ivt mRNA should have, in addition to an optimized 5’ UTR (a CleanCapTM structure followed by an eIF4G aptamer), a tandem repeat of the beta-globin 3’ UTR and be composed of:
a) Adenosine, cytidine, guanosine and uridine residues if the ivt mRNA is to be used in the absence of immune receptors (e.g., in vitro studies in tumour cell lines and production of proteins by tumour cells)
b) Adenosine, cytidine, guanosine and 1-residues if the ivt mRNA is to be used in the presence of immune receptors (e.g., injections in vivo)
Our results presented in Figure 2 differ from previously published results [10-12] that reported that 1-containing mRNA outperforms U-containing mRNA when ivt mRNA is transfected in tumor cells. Differences in the composition (e.g. co-transcriptionally incorporated CleanCapTM and eIF4G aptamer 5’UTR) manufacturing (purification by LiCl precipitation) or transfection (MessengerMax in vitro and TransIT in vivo) may explain this discrepancy. Having shown that the substitution of C residues by 5mC residues reduces the functionality of ivt mRNA (Figure 2), while the substitution of U residues by 1 residues is sufficient to fully abrogate the triggering of immune receptors by ivt mRNA (Figure 3), we recommend that 5mC not be used for the production of ivt mRNA
This work was supported by the University Research Priority Project (URPP) “Translational Cancer Research”, by the Helmut Hornet Stiftung, by the Promedica Stiftung (1406/m and 1412/M), the Krebsliga Schweiz (KFS-4243-08-2017) and by the “MERIT” project supported by the FP7 European Union’s Research and Innovation funding program.


