Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Trieu Tien Sang1, Nguyen Thanh Tung1, Nguyen Thi Thanh Nga1, Ngo Truong Giang1, Nguyen Thi Viet Ha1, Tran Van Khoa1 and Nguyen Thi Trang*2
Received: August 09,2018; Published: August 17,2018
*Corresponding author: Nguyen Thi Trang, Department of Biomedical and Genetics, Hanoi Medical University, N.1, Ton That Tung, Dong Da, Hanoi, Vietnam
DOI: 10.26717/BJSTR.2018.08.001604
Introduction: Spinal muscular atrophy (SMA) is a severe neurodegenerative autosomal recessive disorder. Most of patients are caused by the homozygous absence of a region of exon 7 and exon 8 of the telomeric copy of the SMN gene on chromosome 5. Seting up a molecular diagnostic protocol for detecting SMNt mutation in single cell is basic to Preimplantation Gentic Diagnosis.
Patients and Methods: We test 4 patients and their parent. Lymphocytes of patients and their parent was isolated from fresh blood by ficoll. Taking a lymphocyte by stereoscopic microscope, lysiced the cell, amplifying exon 7 and exon 8 of SMNt gene by using a nested polymerase chain reaction, followed by DraI and DdeI restriction digest of the PCR enabling the important SMNt gene to be distinguished from the centromic SMNc gene which has no clinical phenotype to detect mutation. Electrophoresis PCR products after digesting by restriction enzyme and analysis.
Result: Four patients showed deletion in exon 7, exon 8 SMNt gene. This result is similar with the gene diagnosis from fresh blood.
Conclusion: We have successfully applied the technique of nested-PCR for the gene diagnosis of spinal muscular atrophy from single cell.
Keywords: Spinal Muscular Atrophy; SMN Gene; Nested PCR; Single Cell
Spinal muscular atrophy (SMA) is a severe neurodegenerative autosomal recessive mutation. SMA is characterized by the progressive degeneration of spinal anterior horn cells lead to muscle weakness symmetrical stem limbs, muscle tone and tendon reflex is lost or reduced, chest deformity and stiffness. The worldwide incidence of SMA is 1 / 10,000 live births and the incidence of disease carriers ranges from 1/40-1/60 [1-3]. According to the current international classification, SMA is divided into three categories based on age of disease onset and severity of disease: Severe, intermediate and juvenile form of SMA type I, II, and III [8]3. SMN gene including exon 9 encodes the SMN protein molecules 294 amino acids in length. The SMN gene has two copies, SMNT (SMN1) and SMNc (SMN2) (Figure 1) differ only in 5 base pairs: one in intron 6, one in exon 7, two in exon 7 and one in exon 8. Differences in exon 7 and exon 8 were used to distinguish between SMNT and SMNc in SMA diagnosis [4]. Differ in one nucleotide of exon 7 between SMNT and SMNc, making SMNT synthesize molecules enough length SMN protein and functional, while protein synthesis by SMNc have very limited functionality. There are three types of SMN gene mutations that cause SMA disease: nt 164 Cnt214 y nt 362 nt477 Cnt941 3
a) Type 1: homozygous mutation that lose whole gene SMNt or one part of gene SMNT
b) Type 2: SMNT mutant into SMNc
c) Type 3: Spot mutation occurred in the SMNt gene of one chromosome, SMNt gene on the remaining chromosome in heterozygous pair 5 mutated into form 1 or 2. Approximately 95% of SMA patients with SMNt mutations are in the form of 1 or 2, only in 5% of SMA patients with SMNt mutations in type 3. Both types 1 and 2 are diagnosed. Predicted by the presence or absence of exon 7 SMNt, this is also the basis of the SMA gene diagnostic method (both type 1 and type 2) [5-9].
Treating for these patients creates a burden to both economically and morally for the family and society. Therefore, genetic diagnosis before embryo transfer (preimplantation genetic diagnosis- PGD) SMA families who have a profile with SMA to select healthy embryos transferred to the uterus for implantation mother, from which was born Healthy babies are important and important [10,11]. As the result, we started researching the project: "Establishing assays for detecting SMNt gene mutation in single cell using nested-PCR method". With target: Developed a procedure to identify SMNt gene mutations that cause muscular atrophy from a white blood cell, separated from the peripheral blood of the patient's family.
Pick out 4 families with children aged 0 to 18 years to be examined and treated at Vietnam National Children's Hospital, was diagnosed with spinal muscular atrophy due to loss of exon 7 homozygous gene SMNT
Separating Leukocytes from Peripheral Blood Samples: Leukocyte extraction from whole blood according to the Ficoll- paque protocol. Dilute the white blood cell with 1x PBS solution, pick up a white blood cell on a microscope and place it on a 0.2 ml PCR tube. After that, the cell lysis was 5|iL KOH 0.2M, annealed 65°C for 10 minutes, neutralize KOH with 5|il tricine 0.2 M.
a) PCR: Amplification of exon 7, exon 8 of SMN by nested- PCR technique using two specific primers exon 7, exon 8 SMN gene via two loops: Thermal cycle was performed as follows: 960C, 5 minutes; [940C, 1 minute; 550C, 1 minute; 720C, 1 minutes] x25 cycles; 720C, 10 minutes. Obtain PCR of loop 1 as a template for loop 2 PCR. The PCR cycle was performed on efpendorft amplifier in condition: 960C, 5min; [950C, 30s; 550C, 30s; 720C, 45s] x 35 cycles; 720C, 5min.
b) Cut DNA with Restriction Enzyme DraI and Dde I: PCR products were incubated with restriction enzymes Dra I and Ddel about 2-3 hours. Electrophoresis PCR products with 3% agarose gel. Product after enzyme cutting of exon 7 SMNt gene (188bp) and of exon 7 SMNc gene (164bp and 24bp). Product after enzyme cutting of exon 8 SMNt gene (190bp) and exon 8 SMNc gene (120bp, 70bp).
In Figure 2, exon 7 PCR products were not incubated with DraI (Ko DraI) enzyme, and exon 8 was not incubated with Ddel (Ko DdeI) enzyme from Patient C3’s father (B3), patient C3's mother (M3), C3 patients are the same, with 188bp - product of exon 7 and 190bp - product exon 8. But when incubated with the enzyme DraI PCR products that:
a) B3 and M3 both have 188bp and 164bp respectively, meaning both B3 and C3 have exon 7 SMNT (uncutted- 188bp), exon 7 SMNc is cut into two pieces (164bp and 24bp).
Figure 2: Results of electrophoresis on agarose gel 3% PCR products-patient's family C3: Ladder 50bp.
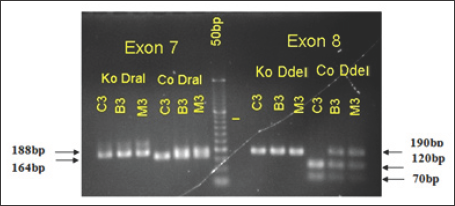
b) Meanwhile the only one C3 has 164bp -loss homozygous exon 7 SMNt, only exon 7 SMNc was cutted by restriction enzymes into two band (164bp, 24bp). This result is consistent with PCR conclusions from the whole sample of Vietnam National Children’s Hospital.
Thus, patients with C3 loss have co-exon 7 and 8 SMNT genes. This illustrate, we were successful in amplified SMN gene from a human leukemia cells and incubated in step restriction enzymes. However, because of the 2% agarose gel electrophoresis results, the two bands of exon 7 SMNc cutted by the enzyme DraI in sample B3, C3 were not clearly separated.
Figure 3: Results of electrophoresis on agarose gel 3% PCR products after incubation with restriction enzyme, patient's family C7: Ladder 100bp.
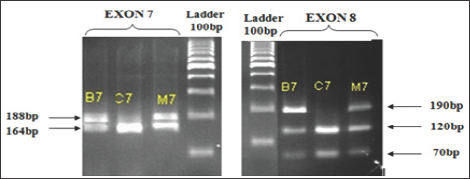
In Figure 3, both father (B7) and mother (M7) of the C7 patient have a SMNt exon 7 gene. Therefore, when incubating the PCR products with restriction enzyme DraI have two-stranded (180bp in exon 7 SMNt and 164bp in exon 7 SMNc), similar with exon 8 of the SMN gene. Thus, patients also lose C7 exon 7 and exon homozygous gene 8 SMNT. Conducting research on other patient families, we obtained the following results: All 4 patients with spinal muscular atrophy research are lost exon 7 SMNt. This result is consistent with the genetic diagnosis SMNt from whole blood samples of Vietnam National Children's Hospital as well as the research of other authors [7-9]. Also, all parents of patients have the exon 7.8 SMNt there and in fact, the parents are non-SMA expression (Table 1). That is, just one chromosome in the pair of homologous 5 gene SMNT normal (heterozygous) can translate enough protein SMN (survival motor neuron) for nerve cells mobilize normal operation.
From the results obtained in the study, we can confirm that the first step we have succeeded in building processes identified gene mutation that causes SMA SMNT from a cell.


