Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Yun Hui Chen1, Tian E Zhang1, Yan Li2, Yong Xiang GAO1, Yuan Yuan GONG1, Chuan Zheng1, Rong Qiang Liu*1 and Wei Huang1
Received: September 14, 2018; Published: October 01, 2018
*Corresponding author: Rong Qiang LIU, College of Basic Medicine, Chengdu University of Traditional Chinese Medicine, Chengdu, Sichuan, China
DOI: 10.26717/BJSTR.2018.09.001810
Introduction: Alzheimer’s disease (AD) is a healthcare and societal priority and the cost of care and the burden on caregivers keep increasing. Danggui-shaoyao-san (DSS), a classic formula of traditional Chinese medicine (TCM), has been widely used in the treatment of AD. Although wellpracticed in clinical medicine, its molecular mechanisms in the treatment of AD remain to be elucidated.
Material and Methods: The candidate compounds of DSS were collected from Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP), the pharmacological indices of each candidate including oral bioavailability and drug-likeness were analyzed, and the protein target of the candidate and corresponding genes were retrieved from TCMSP and UniProt Knowledgebase, respectively. The potential pathways associated with the genes were identified by using the functional annotation tool of The Database for Annotation, Visualization, and Integrated Discovery bioinformatics resource 6.8.
Results: A total of 534 compounds were retrieved from DSS, and 31 active compounds and 58 targets relevant to AD were obtained. Further analysis of gene enrichment acquired 79 significant pathways that interacted with 53 of those AD targets, such as modulating neuroactive ligandreceptor interaction, calcium signaling pathway, TNF signaling pathway and cAMP signaling pathway.
Conclusions: The network pharmacology-based study reveals that DSS’s favorable potency on AD may be associated with its 31 bioactive compounds that interact on 53 targets of AD through 79 significant pathways and may provide a preliminary evidence on how DSS treat AD through multiple components, targets and pathways.
Keywords: Danggui Shaoyao San; Alzheimer’s Diseases; Network Pharmacology
Alzheimer’s disease (AD), a progressive and debilitating neurodegenerative disorder, is the most common cause of dementia with a global prevalence at 46.8 million in 2015. An estimated 5.5 million people in the USA are living with AD in 2017, a number expected to grow to 13.8 million in 2050. By then, nearly one million new cases are projected to develop per year, that is to say one new case develops for every 33 seconds [1]. Treating AD is a healthcare and societal priority and the cost of care and the burden on caregivers keep increasing [2]. However, it is still irremediable yet, and none of the treatments available today prevents or reverses the damage to neurons that causes the disease. Thus, there is an urgent need for more effective therapeutic approaches and researchers are looking for new therapies with breakthroughs. Danggui Shaoyao San (DSS), a classic formula of traditional Chinese medicine (TCM), has become a focus of attention for many researches due to its favorable potency against AD [3,4].
This formula was first recorded in the Jing Gui Yao Lue by Zhongjing Zhang (150–219 A.D.) and its therapeutic effect on AD was first reported by Japan researchers in the 1980s.5 Its protective actions of inhibiting neuronal damage, enhancing cognitive behavior, and improving mitochondrial impairment have been well documented [6-10]. However, DSS’s molecular mechanisms of action on AD are still not fully and clearly elucidated due to the complexity of formula in chemical composition, therapeutic targets, and synergistic interactions among components. The complexity of formula requires an update of the research paradigm from the dominant ‘one target, one drug’ to a new ‘network target, multi-components’ and calls for a method that can decipher the relationships between DSS and AD in a systematical manner. Hence, a set of TCM network pharmacology approach is required to screen the multi-compounds of formula in a high-throughput manner, prioritize disease-associated genes, predict pharmacological actions of herbal compounds and the target profiles, reveal herbal compound-gene-disease co-module associations, and interpret the combinatorial rules and network regulation effects of TCM formula [11].
This approach may provide new ideas and possibilities for elucidating multi-scale action mechanisms of DSS on AD. Herein, this study applied the network pharmacology approach to decipher the DSS’s potency on AD from a systematics perspective and at the molecular level. It could help deepen our understanding of DSS on AD and provide a basis for accelerating the drug discovery of classic TCM formula. The workflow in the present study included four major steps and is illustrated in Figure 1.
DSS consists of six herbs, including Angelicae Sinensis Radix (ASR), Paeoniae Radix Alba (PAR), Poria Cocos (Schw.) Wolf. (PC), Atractylodes Macrocephala Koidz. (AMK), Chuanxiong Rhizoma (CR), and Alisma Orientale (Sam.) Juz. (AO). Its compounds were collected from Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, http://ibts.hkbu.edu.hk/ LSP/tcmsp.php), which consists of 499 Chinese herbs registered in Chinese Pharmacopoeia (2010 edition) with 29 384 ingredients based on critically examined pharmacology and clinical knowledge. Important absorption, distribution, metabolism and excretion (ADME) properties are also provided for further screening and evaluation of ingredients [12].
ADME indexes including oral bioavailability (OB) and Druglikeness (DL) were used to select the ingredients with favorable pharmacokinetics properties from DSS. OB is one of the most vital pharmacokinetic properties of orally administered drugs and plays an important role for the efficiency of the drug delivery to the systemic circulation [13,14]. In this study, the compounds with OB ≥30% were assessed as candidate compounds for further analysis. The DL is a qualitative concept used for estimating the drugability of a substance. Its evaluation helps to screen out excellent compounds and increase the ‘hit rate’ of drug candidates [15]. Herein, the DL of compounds in DSS was calculated using Tanimoto coefficient of [16]:
In which x is the molecular descriptor of DSS, and y represents the average drug-likeness index of all 6511 molecules in the Drugbank database based on Dragon software (http:www.talete. mi.it/products/dragondescription.html). In this study, the active compounds were defined as those with DL indices ≥ 0.15. The compounds meeting OB ≥30% and DL ≥0.15 were treated as candidate compounds for further analysis.
The protein targets of the compounds were retrieved from the TCMSP (http://lsp.nwsuaf.edu.cn/tcmsp.php). The dataset used in model-building comprised 6511 drug molecules and 3987 targets for which the compound-protein interactions are known in the Drugbank database [17]. The gene names were extracted further from the UniProt Knowledgebase (UniProtKB, http:// www.uniprot.org) [18], which is a protein database containing 54,247,468 sequence entries. Subsequently, the obtained targets of the compounds were mapped into the PharmGKB, [19] Drugbank, [17] GAD, [20] OMIM, [21] TCMSP [12] and GEO [22] databases to match the targets related to AD.
The obtained genes were analyzed to further identify the significant pathways targeted by the compounds of DSS using the functional annotation tool of The Database for Annotation, Visualization, and Integrated Discovery Bioinformatics Resource (DAVID v 6.8, http://david.abcc.ncifcrf.gov/). It comprises an integrated online biological knowledge base and analytical tools for systematically extracting biological data from large gene and protein lists [23]. Threshold count more than or equal to 2 and EASE scores less than or equal to 0.05 were selected in functional annotation clustering.
To further explore the mechanisms of actions of DSS on AD, compound–target (C-T) and target-pathway (T-P) networks were generated by using Cytoscape 3.4.0 (http://www. cytoscape. org) [24]. In the graphical network, the ‘nodes’ represent the compounds, protein targets, or signal pathways, and the ‘edges’ encode the interactions between compound and target or between target and pathway. Further, ‘degree’ and ‘node betweenness’ were calculated to measure the topological property of each node in the interaction network. Specifically, the former refers to the number of links to node i; while the latter describes the number of shortest paths between pairs of nodes that run through node i.
A total of 534 molecules were retrieved from TCMSP, including 125 in ASR, 85 in PAR, 34 in PC, 55 in AMK, 189 in CR, and 46 in AO. Among those compounds, 63 compounds met the pre-defined requirements of OB ≥30% and DL ≥0.15 (Table 1). The ferulic acid, ligustilde, Z-ligustilide and 3-Butylidenephthalide, (Z) in ASR and ferulic acid, Z-ligustilide and 3-Butylidenephthalide, (Z) in CR fail to reach the pre-defined requirements, however they have been validated as active compounds in experimental studies with curative effect on AD [25,26]. Hence, they were also included as candidate compounds. Finally, 64 compounds were obtained after removing the duplicates. The details of the obtained compounds were presented in Additional file 2: Table 1.
In total, 43 of 64 compounds in DSS were associated with 448 target proteins. After eliminating the overlapping protein, 120 associated proteins were obtained. Then, the obtained targets were mapped into the PharmGKB, drugbank, GAD, OMIM, TCMSP and GEO. Eventually, 58 targets relevant to AD were reserved for further analysis (Additional file 3: Table 1), and 31 compounds were retrieved after removal of 12 candidate compounds without any relevant targets.
Note: DSS, Danggui Shaoyao San; OB, oral bioavailability; DL, drug-likeness.
The 31 obtained candidate compounds and all their 58 potential targets were applied to generate a network revealing compound-target interactions (Figure 2). This C-T network comprised 89 nodes (31 compounds and 58 targets) and 176 edges (compound-target interactions). It represented a global view of interactions between the potential compounds of DSS (red globe) and targets of AD (gray hexagon). The centralization and heterogeneity of the network were 0.323 and 1.276, respectively. It indicated the compound-target space in the network was biased toward certain compounds and targets. As shown in Figure 2, MOL 000422 (kaempferol) displayed the largest number of interaction with targets (degree = 32), followed by MOL 000358 (beta-sitosterol, degree = 17), MOL 000449 (Stigmasterol, degree = 15), MOL 002111 (3-Butylidenephthalide, (Z)-, degree = 14 ), MOL 002135 (Myricanone, degree = 13), MOL 002122 (Z-ligustilide, degree=12), MOL 000360 (ferulic acid, degree = 11), and MOL 000049 (3β-acetoxyatractylone, degree = 10).
Figure 2: Compound-target network for DSS to treat AD. The gray hexagon nodes represent active compounds, and red globe nodes represent potential target. The edges represent the interaction between them, and the node size is proportional to the degree.
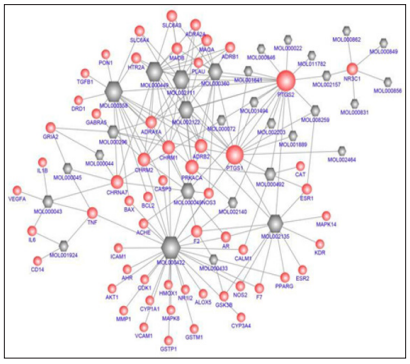
Pharmacologically, the targets with a high degree might play a crucial role in the action of DSS on AD. This C-T network also elucidated that PTGS2 (degree = 21) corresponded to 21 compounds of DSS, followed by PTGS1 (degree = 15), PRKACA (degree = 8), CHRM 2 (degree = 7), CHRM1 (degree = 7), ADRB 2 (degree = 7), CHRNA7 (degree = 7), NR3C1 (degree = 5), F2 (degree = 5), ADRA1A (degree = 5), SLC6A3 (degree =4), MAOB (degree = 4), MAOA (degree = 4), ADRA2A (degree = 4), TNF (degree = 4), ESR1 (degree = 3), ADRB1 (degree = 3), SLC6A4 (degree = 3), HTR2A (degree = 3), NOS3 (degree = 3), AR (degree = 3), and GRIA2 (degree = 3). The remaining 36 targets showed interactions with two or one compounds.
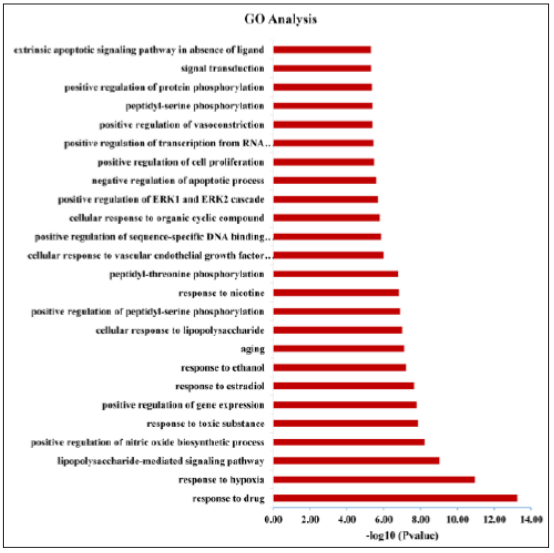
Function of the predicted protein target was investigated using the GO enrichment analysis function of the DAVID V 6.8 platform. A total of 244 entries were confirmed in this analysis. Figure 3 listed the 25 most significantly enriched terms (P < 0.01), and the major biological processes covered signal transduction, cell communication, and energy metabolism. The results revealed the potential targets of DSS and the corresponding genes of AD shared functionally similar characteristics.
Figure 3: Gene analysis of treatment targets. The x-axis represents enrichment analysis ratings (P < 0.01) and the y-axis represents the enrichment analysis of significant types of biological processes.
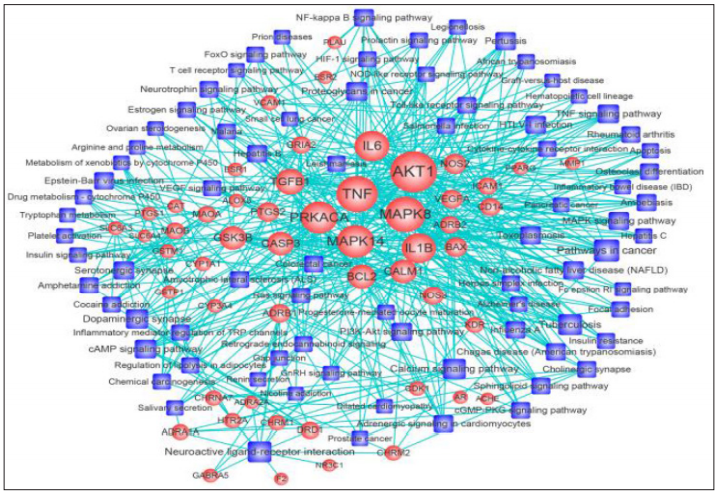
The functional annotation tool of DAVID V 6.8 was used to conduct the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. In total, 79 pathways were significantly associated with the input set of genes (P < 0.05). Then, the T-P network was constructed to further elucidate the underlying mechanisms (Figure 4). This network consisted of 132 nodes (53 targets and 79 pathways) and 517 edges (targetpathway interactions). The centralization and heterogeneity of the network were 0.265 and 0.865, respectively. It indicated that certain nodes were more concentrated in the network than others. The results demonstrated that 45 non-disease pathways out of 79 pathways enriched within multiple targets were essential for the therapeutic effects of DSS on AD, such as Neuroactive ligandreceptor interaction (degree=13), calcium signaling pathway (degree=12), Dopaminergic synapse (degree=11), cAMP signaling pathway (degree=10), TNF signaling pathway (degree=10), PI3K-Akt signaling pathway (degree=9), MAPK signaling pathway(degree=9), Serotonergic synapse (degree=9), Adrenergic signaling in cardiomyocytes (degree=8), NF-kappa B signaling pathway (degree = 8), cGMP-PKG signaling pathway (degree = 7), Osteoclast differentiation (degree = 7), and Neurotrophin signaling pathway (degree = 7). Their interaction information with target proteins were shown in Additional file 4: Table 1.
Figure 4: Target-pathway network of DSS for treating AD. The pink round nodes represent potential targets, and the green square nodes represent significant pathways. The edges represent the interaction between them, and the node size is proportional to their degree.
A TCM formula is a complex system that exerts extensive biological and pharmacological effects through ‘network targets and multi-components’. In view of the complicacy and uncertainty of its data, the approach of network pharmacology has become a necessary complement to the experimental researches for its high throughput. The results of several studies have validated its reliability, efficacy and perspectiveness [27-31]. Herein, instead of following the dominant ‘one target, one component’ paradigm, we adopted the approach of network pharmacology that focused on the concept of ‘network target, multicomponent therapeutics’ with a holistic view shared by TCM. In the present study, 31 bioactive compounds from DSS interacted with 53 targets through 79 significant pathways. It might provide a preliminary evidence on how DSS treat AD through multiple components, targets and pathways in a systematic view. Meanwhile, it also provides new perspectives to identify multi-scale mechanisms of action of TCM formula to treat diseases. As an empirical system of multicomponent therapeutic, TCM formula exerts extensive biological and pharmacological effects through multiple compounds and targets.
In the network pharmacology, the degree of node is a key topological parameter that characterizes the most influential nodes, and those high-degree nodes with more compound-target interactions are likely to play more important roles. In the present study, network analysis suggested that each target could interact with multiple molecules (3.05 compounds per target on average), and one compound also acted on several targets (5.53 targets per compound on average) synergistically to treat AD. More than two thirds (24 of 31) of compounds were linked with greater than or equal to two targets, indicating the ‘multi-components, network target’ properties of DSS’s mechanism of action on AD. A further analysis on the 177 C-T interactions revealed that 7 out of 31 compounds were with high-degree distribution, and each hit more than 10 potential targets. Among which, 5 compounds were from ASR (i.e. beta-sitosterol, Stigmasterol, 3-Butylidenephthalide, (Z)-, Z-ligustilide, and ferulic acid), 4 from CR (i.e. 3-Butylidenephthalide, (Z)-, Myricanone, Z-ligustilide, and ferulic acid), and 2 from PAR (i.e. kaempferol and beta-sitosterol).
In general, the higher the degree, the more pharmacologically important the compound, indicating that ASR, CR and PRA might have more pharmacological effects on AD than other three herbs of DSS in the C-T network.
Notably, among the 31 compounds, kaempferol from PAR exhibited the largest number of interaction with AD targets (32 of 177 edges, 18.1%), followed by beta-sitosterol from PAR and ASR (17, 9.6%), Stigmasterol from ASR (15, 8.5%), and 3-Butylidenephthalide, (Z)- from ASR and CR (14, 7.9%). These compounds were reported to possess multiple pharmacological actions. For instance, kaempferol was proven capable of inhibiting function of enzymes involved in enhancing expression and/or activity of several anti-oxidant enzymes to protect neural cells from DNA damage, lipid peroxidation and induction of apoptosis or cell death [32]. Among 58 proteins, PTGS2 possessed the largest number of connected compounds, followed by PTGS 1, PRKACA, and CHRM2, indicating many proteins targeted by DSS might display similar binding patterns with the ligands. PTGS2, as a proinflammatory agent, has been found to exacerbated inflammation and to induce extensive neuronal death and apoptosis [33].
To better understand the action mechanism of DSS for AD therapy, the potential targets were retrieved on KEGG database to extract the canonical pathways, and then the T-P network was built. The results suggested nearly two thirds of the targets (34 out of 53) located in multiple pathways (n ≥ 4), whereas 16 nondisease pathways were intensively connected to the targets(n ≥ 7), including neuroactive ligand-receptor interaction, calcium signaling pathway, TNF signaling pathway, cAMP signaling pathway, PI3K-Akt signaling pathway, MAPK signaling pathway, NF-kappa B signaling pathway, Adrenergic signaling in cardiomyocytes, Toll-like receptor signaling pathway, Sphingolipid signaling pathway, Neurotrophin signaling pathway, and cGMP-PKG signaling pathway. It indicated that DSS might exert its therapeutic effects on AD through multiple pathways. Remarkably, neuroactive ligand-receptor interaction was regulated by 13 potential targets in the T-P network and should be regarded as the crucial pathway.
It has been well testified to modulate the neurotransmitters including glutamate, serotonin, cholinergic receptor, and histamine receptor, which play an important role in cognition and memory [34]. Our study also consisted with the calcium hypothesis that the disruption of calcium signaling pathway underlies the pathogenesis of AD. Calcium ions (Ca2+) are essential second messengers that regulate a wide variety of important eukaryotic cells functions such as differentiation, proliferation, growth, survival, apoptosis, gene transcription and membrane excitability and the restoration of calcium homeostasis has been regarded a promising strategy for the treatment of AD [35-37]. In addition, the effects of TNF in such neurodegenerative disorders as AD is getting increased attention. It involves in AD-related brain neuro inflammation and contributes to amyloidogenesis via β-secretase regulation, implying its potency for future AD therapy [38].
The present network-pharmacology based study provides a holistic understanding of the underlying pharmacological mechanisms of DSS in the treatment of AD. It reveals that DSS’s favorable potency on AD may be associated with its 31 bioactive compounds that interact on 53 targets of AD via 79 significant pathways, such as modulating neuroactive ligand-receptor interaction, calcium signaling pathway and TNF pathway. This study provides a preliminary evidence on how DSS treat AD in a systematic view and may facilitate the drug discovery process of TCM formula for the treatment of AD. Our future research will aim to provide more experimental insights into this aspect.
This study was supported by the National Natural Science Foundation of China (No.: 81603537); International Cooperation and Exchange Project of Sichuan Provincial Science and Technology Department (No.: 2017HH0004) and Key Project of Sichuan Provincial Education Department (No.: 17ZA0163).


