Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Yingjun Xie1, Wei Jian2, Jingsi Chen2, Dunjin Chen2 and Xiaofang Sun*1
Received: October 01, 2018; Published: October 11, 2018
*Corresponding author: Xiaofang Sun, Key Laboratory for Major Obstetric Diseases of Guangdong Province, Key Laboratory of Reproduction and Genetics of Guangdong Higher Education Institutes, The Third Affiliated Hospital of Guangzhou Medical University, Duobao Road 63#, 510150, Guangzhou, China
DOI: 10.26717/BJSTR.2018.09.001876
The 7q36 microdeletion has been identified in patients with variant phenotypes including sacral agenesis, holoprosencephaly and intellectual disability. Here we describe a case of fetus with hemivertebrae and scoliosis and detected a 6.42 Mb pure microdeletion at 7q36.1-qter by chromosomal microarray analysis (CMA) that was not determined by traditional karyotyping. This microdeletion was confirmed by Fluorescent in situ hybridization (FISH) assay. Accurate breakpoints of the deletion in this case were used to establish correlations between microdeletion at 7q36.1-q36.3 and the accompanied phenotypes, hemivertebrae deformity, which is rarely found in monosomy 7q36.1-qter. Our study identified and described an important relationship between fetal hemivertebrae with scoliosis and 7q36.1-qter microdeletion overlap with MNXI and SHH.
Abbreviations: CMA: Chromosomal Microarray Analysis; FISH: Fluorescent In Situ Hybridization; CS: Congenital Scoliosis; HPE: Holoprosencephaly; SMMCI: Solitary Median Maxillary Central Incisor
Congenital scoliosis (CS), a lateral curvature of the spine resulted from vertebral defects leading to a failure of vertebral formation and vertebral segmentation, occurs in about 1 per 1,000 live births, might be caused by variant factors such as environmental changes, genetic variations or both [1]. The major forms of vertebral defects include block vertebrae, hemivertebrae, butterfly and wedged vertebra, and unsegmented bars. Hemivertebrae usually represents an extra vertebral segment which is due to complete failure of vertebral formation and is the most common cause of CS. Vertebral malformations that result in CS may be linked with genetic syndromes such as Alagille syndrome, spondylocostal dysostosis, and Jarcho-Levin syndrome [2]. However, until now, the etiology of congenital scoliosis remained largely unknown [1]. In our case, we detected a 6.42Mb pure deletion in 7q36.1- qter (Chr7:150,915,999- qter) using chromosomal microarray analysis (CMA) in uncultured amniocytes derived from the mother of fetus with hemivertebrae and scoliosis. Several OMIM genes, including KCNH2(152427), NOS3(163729), PRKAG2(602743), DPP6(126141), PAXIP1(608254), EN2(131310), NCAPG2(54892), RHEB(601293), SHH(600725) located in this genomic region. Indeed hemivertebrae has rarely been reported in patients carrying pure 7q36.1-qter microdeletion, especially the prenatal diagnosis.So our study identified and described an important relationship between fetal hemivertebrae with scoliosis and 7q36.1-qter microdeletion.
Clinic Data: A 29-year-old primiparous woman at 22 weeks of gestation was referred to the prenatal diagnosis clinic for genetic consultation because of fetal congenital scoliosis, such as fetal spinal segmental T10 - T12 to the right angle of bent into a triangle, stenosis. Spine coronary section showed that the right period of T10 - T12 thoracic vertebral ossification center was absent ,which was detected by gestation prenatal ultrasound at 22 weeks. Her husband was 30 years old and this couple was non-consanguineous without family history of congenital malformations on either side. This pregnant woman at 12 weeks repeatedly appeared a small amount of vaginal bleeding and experienced long-term tocolytic treatment. Amniocentesis was performed at 24 weeks of gestation. Because of the severe malformations of the hemivertebrae deformity, abortion was suggested. The pregnancy was subsequently terminated with a female fetus visually normal. Organ malformations could not be examined as the parents did not consent to a fetal autopsy. All data was collected with the informed consent of the patients.
Cytogenetic Analysis: With informed consent, amniocytes (15ml) was collected by amniocentesis, and the sample was subjected to amniocyte culture according to a standard cytogenetic protocol. Meanwhile, 5ml of peripheral blood was collected from each parent. The blood samples were subjected to lymphocyte culture according to the instructions of the standard blood cytogenetic protocol. Finally, the cultured amniocytes and lymphocytes were assayed with conventional cytogenetic analysis using Giemsa-banding methods with a resolution of 550 bands. We also carried out FISH using the chromosome 7qer probe and LSI 7q11.2 probe (Vysis, USA) to exclude low-level mosaicism (600 nucleus have been analyzed), according to the manufacturer's instructions.
MA Analysis: Amniocytes (10ml) was collected from the pregnant woman at 24 weeks of gestation by amniocentesis. The peripheral blood specimens were from the parents. Genomic DNA was extracted from the uncultured amniocytes using a QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, D). The genomic DNA (250 ng) was amplified, labeled and hybridized to the Agilent human CGH microarray (Agilent Technologies, CA) containing unique 60-mer oligonucleotides representing 244,000 features with an average probe spatial resolution of 6.4Kb. Labeling reactions were performed with 1mg purified genomic DNA and Agilent Genomic DNA Labeling Kit PLUS (Agilent, CA) according to the manufacturer's instructions. Labeled samples were hybridized into the microarray chip using Agilent Oligo array CGH Hybridization Kit, the chip was then secured in the Hybridization Chamber and incubated at 65 °C for 40h in the Agilent Hybridization Owen Rotator (20rpm). Data analysis was performed using the Agilent Feature Extraction 9.1 and CGH Analytics 3.5. In brief, a log2 expression ratio was computed and normalized for forward and reverse fluorescent dye (i.e., dye-swap) experiments using the CGH Analytics 3.5 Software.
Normal karyotypes of 46 chromosomes were detected in the peripheral blood of both parents (data not shown) and in the cultured amniocytes (Figure 1). The inability to find such a microdeletion with conventional cytogenetic techniques might be owing to the insufficient resolution of G-banding. We further performed CMA analysis in the cultured amniocytes and found a 7q36.1-qter (chr7: 150,915, 999-qter) microdeletion of ~6.4Mb in the patient (Figure 2). Then FISH analysis was performed to explore gross chromosomal aberrations and showed that the couples had intact 7q telomeres (Figure 3a & 3b) while a de novo distal 7qter microdeletion was detected in the cultured amniocytes (Figure 3c).
Figure 1: Karyotyping of the cultured amniocytes. The karyotype was normal in the amniocytes when the chromosomes were visualized using G-banding techniques with a resolution of 550 bands.
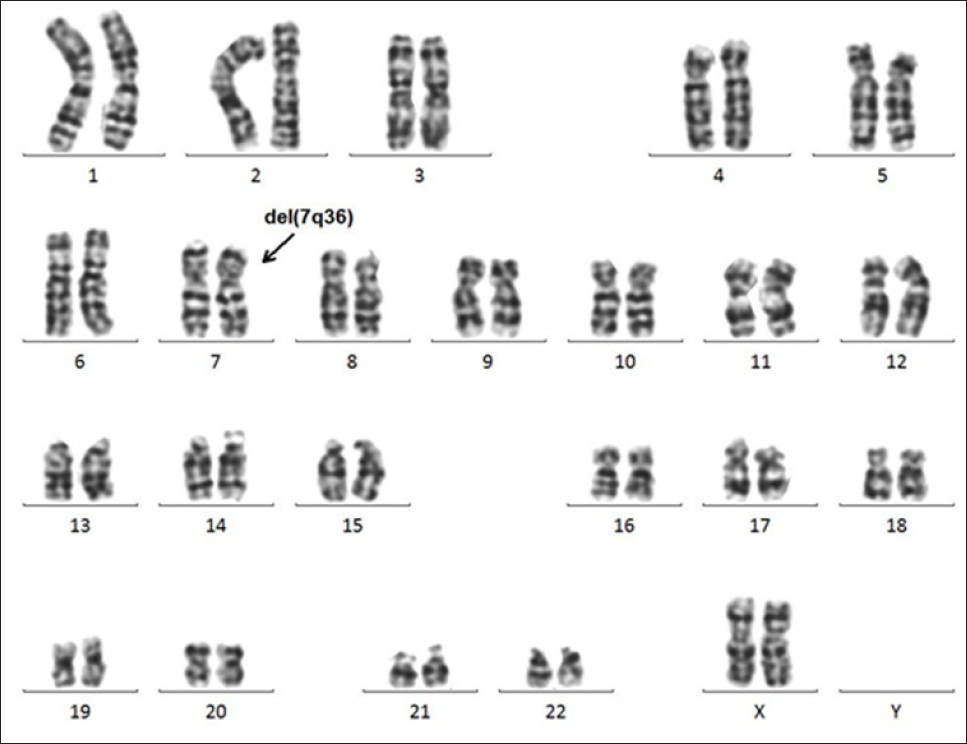
Figure 2: Metaphase fluorescence in situ hybridization (FISH) analysis of the cultured lymphocytes and amniocytes using CEP7 probes and 7q subtelomere-specific probes. No distal deletion was detected on 7qter in the cultured lymphocytes that were derived from the father .
Note:
a) Or the mother
b) Indicated by white arrowheads. The lack of a 7-qter signal on one chromosome 7 revealed the monosomy 7qter in the amniocytes
c) Indicated with orange arrowhead.
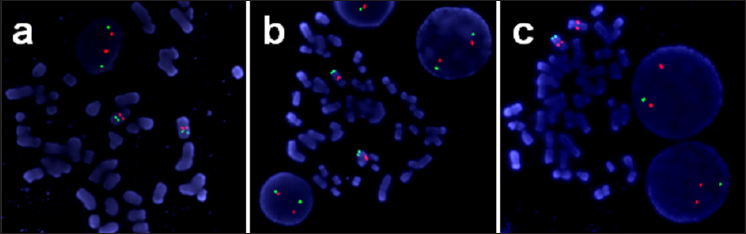
Figure 3: Microdeletion at 7q36.1-qter. The microarray profile of the 7q36.1qter microdeletion is represented by the bold red line. The schematic diagram is shown below, and the deleted region is outlined with a red box.
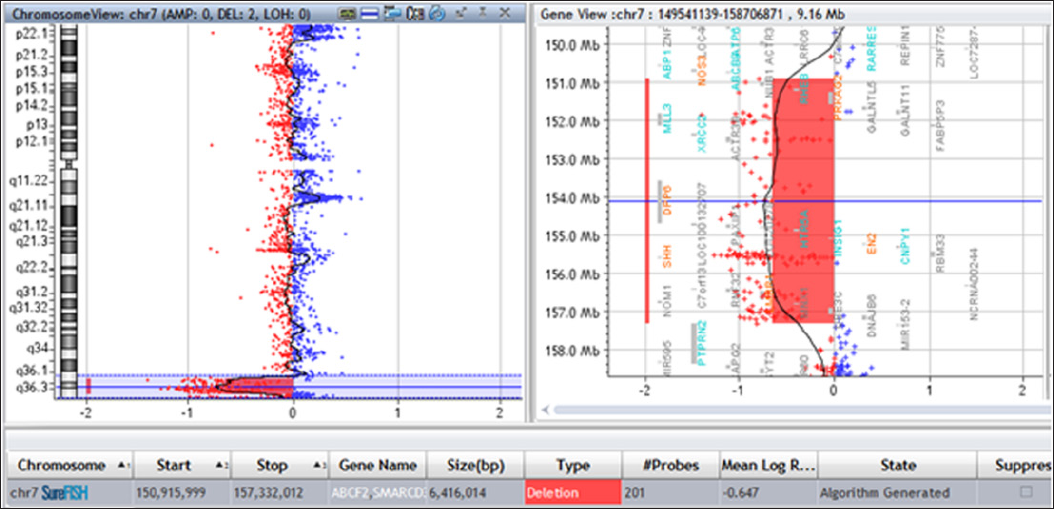
Diagnosis basing on clinical symptoms of the deletion of the long arm of chromosome 7 (7qter) is more difficult than most other chromosomal abnormalities due to the wide related phenotype variations. Currently terminal deletions of 7 qter are well known and are frequently linked with holoprosencephaly or hypotelorism due to the involvement of the SHH gene located in 7q36.3 [3-8]. The typical clinical phenotypes of 7qter include congenital limb deformity (polysyndactyly), brain malformations, often with the holoprosencephaly spectrum, mental retardation and short stature [4,6,7,9]. Here we revealed and described a fetal patient carrying hemivertebrae with scoliosis by prenatal ultrasound, and discovered a 7q36.1-qter (chr7: 150, 915, 999-qter) microdeletion of ~6.4Mb. This phenotype was rarely reported previously in patients carrying 7qter abnormalities. We further compared the clinical phenotypes of the fetus patient, with other reported patients who carried 7q36.1-qter deletions, who were reported to have the hemivertebrae or scoliosis which is similar to our patient (Table 1). Lee C.G reported a patient who carried a deletion of 6.89-Mb on 7q36.1-q36.3 (chr7:148,751,906-155,637,739) and suffered moderate scoliosis, microform holoprosencephaly with microcephaly, distinctive facial features, severe intellectual disabilities, behavior problems, seizures, short stature, penoscrotal transposition, and ulnar ray deficiency [7]. Coutton et al. reported a 7q36.3 deletion of ~2.7Mb (chr7: 155,686,857-158,384,574, NCBI, hg19) in a patient with Currarino Syndrome and Holoprosencephaly (HPE) microform with abnormalities between T8 and T10 vertebras [4]. Fruhmesser et al. identified a 7q36.2-qter deletion of about 4.4Mb in a patient with scoliosis phenotype [6]. Here our patient carried a pure deletion of 7q36.1-36.3 and suffered from hemivertebrae.
Therefore, our findings provided important evidence to elucidate the important role of 7q36 in vertebral development. By comparation of the genomic deletions in these patients, we noticed that 7q36.3 region was encompassed by all the deletions in patients showing hemivertebrae pathogenesis or scoliosis. So our results provided important information for the mechanisms underlying scoliosis and vertebral development. The ~6.4 Mb region deleted in our case contains at least 27 protein coding genes, in which 24 genes with OMIM annotations, including EN2(MIM 131310), SHH(MIM 600725), PRKAG2(MIM 602743),DPP6(MIM 126141), and MNX1(MIM 142994). Though so many genes located in 7q36 region, the MNXI gene was quite important as it has been implicated in sacral development. Indeed, mutations in MNXI (chr7:156,786,745-156,803,345) have been identified in dominantly inherited sacral agenesis [10] and were associated with Currarino syndrome, which is also known as caudal regression syndrome or hereditary sacral agenesis. The lowest sarcral bones in these patients at the base of the spine and the coccyx (the lowest element of the backbone) fail to form properly [11]. And a patient who carried a 6.89-Mb sized deletion on 7q36.1-q36.3 (chr7:148,751,906-155,637,739) suffered moderate scoliosis [7], while our patient showing scoliosis carried a 7q36.1-qter (chr7: 150, 915, 999-qter) microdeletion.
We noticed that the overlapping region between the two patients encompassed PRKAG2, DPP6, PAXIPI, EN2, and SHH. In addition, deletions on chromosomes 7q36 involving holoprosencephaly (HPE) spectrum overlap with SHH are association with Solitary median maxillary central incisor (SMMCI) syndrome [12]. As a result, SHH seems to play an important role in the malformation in our patient. A protein, encoding by SHH gene, is instrumental in patterning the early embryo. It has been regareded as the pivotal inductive signal in patterning of the ventral neural tube, the anterior-posterior limb axis, and the ventral somites [13-15]. Furthermore, MNXI and SHH both carry high HI scores of 0.84 and 0.66, respectively, indicating these two genes are more likely to exhibit haploinsufficiency [16]. These considerations suggest that MNXI and SHH deletion may contribute to the CS phenotype observed in our patient. With the improvements of cytogenetic techniques, more and more phenotype-genotype correlations have been discovered. In our study, we used CMA and FISH assay to unravel an association between 7q36 monosomy and serial anomalies, including hemivertebrae with scoliosis. Indeed, previous studies have shown that phenotypes of different patients the 7q36 deletion showed difference, our reports about the correlation between 7q36.1-qter and hemivertebrae/scoliosis expanded our understanding about defects associated with 7q36.1-qter deletion. Further analysis to narrow down the causal region in 7q36.1-qter would be important for prenatal diagnosis and disease mechanism studies.
We are grateful to the patient and his parents for their participation in this study. We would like to thank the staff of the prenatal center for patient's data.
This study was supported by the Natural Science Foundation Committee of China (NSFC-81402652).


