Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Umamaheswari Natarajan1,2, Thiagarajan Venkatesan2, Ali Alaseem2-4, Vijayaraghavan Radhakrishnan', Shila Samuel1 and Appu Rathinavelu*2,4
Received: October 12, 2018; Published: October 29, 2018
*Corresponding author: Appu Rathinavelu, Rumbaugh-Goodwin Institute for Cancer Research, Nova South eastern University, USA
DOI: 10.26717/BJSTR.2018.10.001963
Histone Deacetylases (HDACs) are evolving as key enzymes in many physiological processes, including chromatin remodeling, genome stability, DNA repair, regulation of transcription metabolism, protein secretion and cell cycle progression. In the cellular systems there is a balance between deacetylation and acetylation which controls gene transcription. The HDACs catalyse the elimination of the acetyl moiety from the lysine residues of proteins, as well as the core nucleosomal histones. Through removal of critical acetyl groups from the histones, HDACs can generate a chromatin conformation that can prevent the transcription of genes that encode for proteins involved in cell cycle regulation. In many cancer cell lines, overexpression or activation of the HDAC enzymes results in histone hypo-acetylation and following promotion of pro-cancerous mechanisms. Therefore, HDAC inhibitors represent a potential new class of anti-tumor agents with cytotoxic activity and the ability to regulate gene expression in tumor cells. In the present study, we evaluated the effects of SAHA (Suberoylanilide Hydroxamic Acid), which is a potent inhibitor of HDAC, on cell cycle regulation in LNCaP-MST cells (MDM2 overexpressing prostate cancer cells) and MCF-7 (MDM2 expressing breast cancer cells).
The gene expressions analyzed through qRT-PCR, RT-PCR and western blot of LNCaP-MST and MCF-7 cells, after treating with SAHA (7.5 μm), were able to trigger p21 protein expression. In this study, SAHA seems to be inducing the p21 expression in a p53 independent manner. Our results confirm that SAHA treatments could significantly up and down-regulate some of the key genes and proteins such as p21 that modulate cancer cell growth. In the LNCaP-MST cells, AURKB (Aurora Kinase B), CDC25C (Cell Division Cycle 25C), and CDK1 (Cyclin-Dependent Kinase 1) protein expressions were down-regulated, which may impact the G1 or G2/M phase cell cycle arrest and suppression of cancer growth. Our results with LNCaP-MST cells offer convincing evidence to suggest that the inhibition of HDAC can control proliferative cell signals.
Breast cancer is one of the most common malignant disease in women it has been estimated that, in the USA about one in eight women (about 12.4%) will develop invasive breast cancer, there will be an estimated 266,120 new cases of invasive breast cancer diagnosed in women and an estimated 41,400 breast cancer deaths (40,920 women, 480 men) will occur in 2018 [1]. Although breast cancer rates have been increasing gradually over the past 30 years, the deaths caused by this cancer are on the decline, due to the advancements that have been achieved in early diagnosis and successfully treating the cancer with new therapeutic strategies [2]. On the other hand, prostate cancer is the second-most common cancer diagnosed among men in the USA, [3] resulting in over 164,690 estimated new cases and 29,430 deaths [1]. Due to the advent of the Hallmarks of malignancy, new opportunities for therapeutic targeting are constantly explored [4]. Some of the rekindled approaches include triggering of apoptosis, necrosis, induction of senescence, abrogation of angiogenesis, inhibition of tumor invasion and metastasis.
For achieving the outcomes listed above, new compounds that are designed to impact these hallmarks as a monotherapy or in combination with existing cytotoxic treatment or immunotherapeutics are constantly being developed. Novel agents are required to overcome many of the existing hurdles, which contain drug resistance, deficiency of target receptor expression in tumors, excessive toxicity and as a result comparatively small improvements in survival [5-8]. One of the newly tested drugs is SAHA (Suberoyl Hydroxamic Acid) which belongs to Class I, II and IV of HDAC inhibitors that is effective against multiple types of cancer cells [9-12]. Histone Acetyltransferases (HATs) and HDACs are important components that disturb the dynamics of chromatin folding during gene transcription [13]. HDACs are influence epigenetic modifications in the regulation of several cell signalings, as their inhibition potentiates the therapeutic efficacy of anticancer agents [14-17]. SAHA is a pan HDAC inhibitor that reduces the activity by acting on all 11 human HDACs that belong to Class I, II and IV [18].
As a result of their specificity, SAHA drastically changes cellular acetylation patterns and causes cell cycle arrest in both in vitro and in vivo tumor models [18,19]. Typically, HDACs modify the histone acetylation by catalyzing the removal of acetyl groups from the NH2- terminal lysine residues of the core nucleosomal histones. Such modulation of the acetylation status is involved in the regulation of the transcriptional activity of certain genes. Modifications in both HAT and HDAC activity have been reported to occur in various cancers [9] which were linked to the covalent modification of histones and significant changes in chromatin architecture and gene expressions related to cancer progression [20,21]. Therefore, aberrant exertion of HDAC activity has been found to be concomitant with the development of human cancers [22]. In addition to imparting epigenetic alterations, HDACs regulate the level of protein acetylation, along with Histone Acetyltransferases (HATs). Therefore, our study presented here focused on the impact of SAHA treatment on gene expressions that are critical for cell cycle arrest and cell survival in LNCaP-MST and MCF-7 cells. In order to strongly validate the mechanistic impact of SAHA, we analyzed the expression levels of key genes related to the cell cycle pathway using qRT-PCR, RT-PCR and western blot analysis techniques. Our experiments results show enhancement of cell cycle arrest by SAHA treatments in both LNCaP-MST and MCF-7 cells.
The LNCaP-MST and MCF-7 cells were grown in Dulbecco's Modified Eagle's Medium (DMEM). The growth media for both cell lines were supplemented with 10% fetal bovine serum, 1% Amphotericin B and Penicillin-Streptomycin. Cells were cultured in a humidified atmosphere with 95% air and 5% CO2. When LNCaP- MST and MCF-7 cells reached 75-80% confluency, both cell lines were treated with 7.5 μM SAHA for 24 hrs and then used for RNA and protein extraction.
The cell viability was determined to assess the effect of SAHA treatments using the MTT assay, a colorimetric assay for estimating mammalian cell viability based on the ability of viable cells to reduce yellow 3-(4, 5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) by mitochondrial succinate dehydrogenase. The MCF-7 and LNCaP-MST cells were plated at a density of 10 x104 cells/well in 96-well plates, and incubated at 37oC under 5% CO2 for 24 hrs. Then, the cells were exposed to different concentrations such as 0.5, 2.5, 5.0, 7.5 and 10.0 μM of the test compound (SAHA) for 24 hrs. After which, 10 μL of MTT reagent was added to each well. The plates were further incubated at 37°C and 5% CO2 for 4 hrs in the dark. At the end of incubation 100 μL of DMSO was added to the each well and the absorbance (OD) was read at 490 nm using a Multiskan microplate reader (Molecular Devices Inc., Sunnyvale, CA, USA).
Total RNA was extracted from LNCaP-MST and MCF-7 (control, SAHA treated) cells. The RNA separation was performed using the RNeasy mini-kit according to manufacturer's protocol (Qiagen, Valencia, CA, USA). The quality and concentration of RNA was analyzed by measuring the ratio of absorbance at 260/280 nm.
Alterations in the gene expression of certain genes such as p21, MDM2, CDC25A, CDC25C, AURKB and BIRC5 were analyzed in LNCaP-MST and MCF-7 cells. The gene expressions were compared between control and treatment groups. While analyzing the data for gene expression the β-actin values used for normalization. The qRT-PCR reaction volume of 20 μL contained 50 ng mRNA, 0.4 μM forward and reverse primers, 10 μL of 2x SensiFAST SYBR Hi-ROX One-Step Mix, 0.2 μL Reverse transcriptase and 0.4 μL Ribosafe RNase inhibitors (10U/μL) (Bioline, Taunton, MA, USA). The primer sequences used for qRT-PCR are given in the Table 1. The qRT-PCR reaction was started with reverse transcription step at 45°C for 10 min, followed by cDNA was amplified in 40 cycles with the denaturation at 95°C for 5 sec, annealing at 60°C for 10 sec and extension at 72°C for 5 sec. All reactions were performed in triplicates on the ABI Step One Plus Real-time PCR instrument (Applied Biosystems, Foster City, CA, USA).
For the purpose of validating the changes in the genes that were significantly altered, the RNA was isolated as described previously and then it was reverse transcribed using the one-step reaction and then by using the Access RT-PCR system (Promega, Madison, WI, USA) that contained and optimized buffer, AMV reverse transcriptase for the first-strand synthesis and Tfl DNA polymerase for the synthesis of cDNA. Amplification of specific genes such as p21, p53, AURKB, CDC25A, CDC25C, CDK1, MDM2, BIRC5 and β-actin were accomplished using specific primers as per manufacturer's protocol. The RT-PCR reaction products were separated on 1.5% agarose gel containing fluorescent DNA dye (VWR life sciences, USA). The DNA images were captured using a Bioimaging system (UVP, Upland, CA, USA). The quantitative comparison of each DNA band was performed by measuring band intensity using the ImageJ program (NIH Image, Bethesda, MD).
The LNCaP-MST and MCF-7 control cells, and 7.5 μm SAHA treated cells were lysed with Radio-Immunoprecipitation Assay (RIPA) lysis buffer containing sodium orthovanadate and protease inhibitor cocktail (Santa Cruz Inc., Dallas, TX, USA). Cell lysates were clarified by centrifugation at 4°C for 20 min at 14,000 rpm, and protein concentrations were quantitated using the bicinchonic acid protein assay method (Thermo Fisher Scientific, Grand Island, NY, USA). For the western blot analysis, equal concentrations of protein were separated using sodium dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and blotted onto a nitrocellulose membrane (GE Healthcare, Pittsburgh, PA, USA). Membranes were blocked using proteins from non-fat dry milk and BSA, it probed with specific antibodies for p21, p53, p-p53, AURKB, CDC25C, CDK1, survivin (Cell Signaling Technologies, Danvers, MA, USA), MDM2 (Santa Cruz Inc., Dallas, TX, USA) and β-actin (Sigma- Aldrich, Saint Louis, MO, USA). In conclusion, for the detection of specific proteins, the membranes were incubated in a LumiGLO Reserve Chemiluminescent substrate solution (KPL, Gaithersburg, MA, USA). Densitometric quantification were accomplished using the ImageJ software (NIH Image, Bethesda, MD).
The data presented in the manuscript are the mean + SD from Statistical significance between the control and treated groups was analyzed by unpaired t test. P< 0.05; P< 0.01 were considered statistically significant.
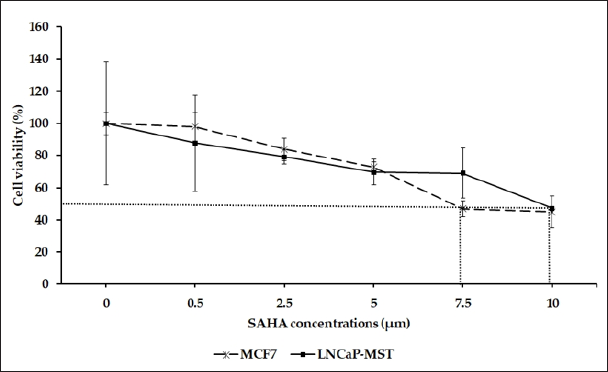
The effect of SAHA on the cell viability of the LNCaP-MST and MCF-7 cells were determined by MTT assay (Figure 1). SAHA treatment inhibited cell viability in a dose- and time-dependent manner in both cell lines. As shown in Figure 1, Both LNCaP-MST and MCF-7 cells sensitive to SAHA treatment, showing a decrease in cell viability at all doses in 24 hrs treatments.
Figure 1: Assessment of cell viability using LNCaP-MST and MCF-7 Cells after treating with SAHA. The effect of 24 hrs treatment on above mentioned cell viability were assessed using 0.5, 2.5, 5.0, 7.5, 10.0 μM concentrations of SAHA. The data are presented as means ± S.E.M. from minimum of 3 independent experiments.
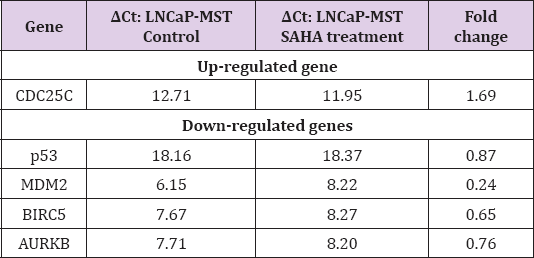
The qRT-PCR results confirmed the changes in expression levels of CDC25A, CDC25C, p53, MDM2, BIRC5 and AURKB, in LNCaP-MST and MCF-7 cells after SAHA treatment as shown in Tables 2 & 3. In LNCaP-MST cells, CDC25A and BIRC5 genes were down-regulated compared to the untreated cells (Table 2). In addition, in the MCF-7 cells, MDM2 and BIRC5 levels were down-regulated more significantly than others (Table 3).
Table 2: Analysis of qRT-PCR for certain genes that are up- and down-regulated in LNCaP-MST cells after SAHA treatment.
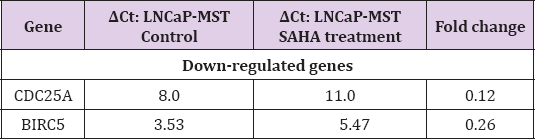
Table 3: Analysis of qRT-PCR for certain genes that are up- and down-regulated in MCF-7 cells after SAHA treatment.
To further confirm the results obtained from qRT-PCR experiments, the RT-PCR was also carried out long template primers. In support of the results of our qRT-PCR experiments, Figures 2&3 show the expression levels of the above-mentioned genes that were considerably altered in LNCaP-MST and MCF-7 cells. The different levels of RNA expressions found in the cells are reflected in the band intensities of RT-PCR. In LNCaP-MST cells p21 was up-regulated by 135%. Interestingly, in MCF-7 cells also the p21 was up-regulated by 161% while the BIRC5 was down-regulated significantly near to complete knockdown levels. On the other hand, the mRNA levels of AURKB, BIRC5, CDC25A, were down-regulated by 23%, 41%, 34% respectively in LNCaP-MST cells. While the mRNA levels of CDC25A was not altered in MCF-7 cells after the SAHA treatment.
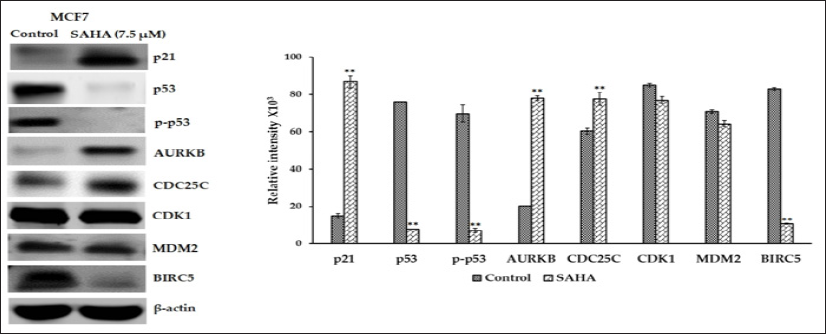
The protein expressions of selected genes were determined in the LNCaP-MST and MCF-7 cells after SAHA treatment. In LNCaP- MST cells, protein expression of Aurora Kinase B, phospho-p53, CDC25C, Survivin, MDM2 and CDK1 levels were significantly down-regulated after 7.5 μm SAHA treatments and the level of p21 was significantly up-regulated compared to the control (Figure 4). In MCF-7 cells, the protein levels such as p21, CDC25C and Aurora Kinase B were up-regulated and p53, phospho-p53 and Survivin expressions were down-regulated after SAHA treatment (Figure 5). The cell cycle phosphatase CDC25C is another cell cycle regulator involved in the transition from the G2 phase to M in the cell cycle regulation [23]. In the LNCaP-MST cells, CDC25C levels were significantly down-regulated after 7.5 μm SAHA treatment, though the mRNA levels were not altered. The CDK1 protein levels were also not altered by SAHA treatment in the MCF-7 cells. For some reason the mRNA expression levels of AURKB (MCF-7 cells), and CDC25C (LNCaP-MST cells) are not correlated to the protein expressions. This may be due to the degradation and stabilization of the proteins; however, further studies are needed to elucidate the exact mechanism that may responsible for this disparity observed in MCF-7 cells.
Figure 2: RT-PCR pictures illustrates that p21, p53, AURKB, CDC25A, CDC25C, CDK1, MDM2, BIRC5, and β-actin mRNA levels in LNCaP-MST cancer cells after SAHA treatment (7.5 μm). Results were statistically analyzed with an unpaired t-test. *: P < 0.05; **: P < 0.01.
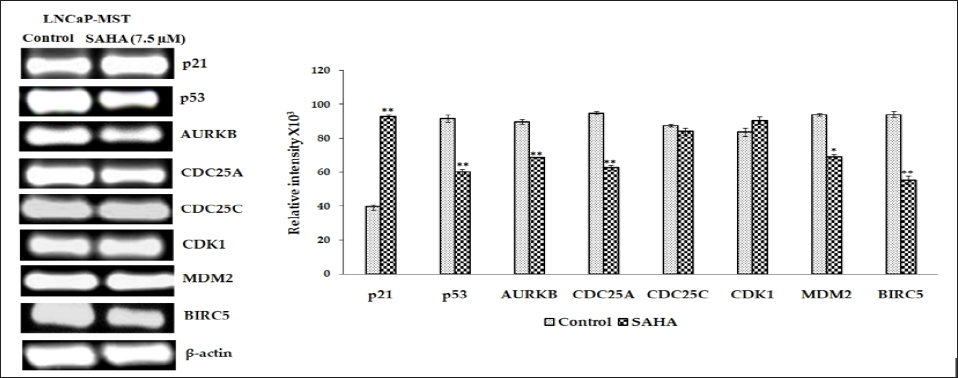
Figure 3: RT-PCR pictures showing p21, p53, AURKB, CDC25A, CDC25C, CDK1, MDM2, BIRC5, and 3-actin mRNA expression levels in MCF-7 cancer cells after SAHA treatment (7.5 μm) and bar graph showing the relative intensity of bands corresponding to differentially expressed genes analyzed using Image J program. Results were analyzed with an unpaired t-test. **: P < 0.01. LNC.IP-MST Control SAHA (7.5 mM) k p21 120
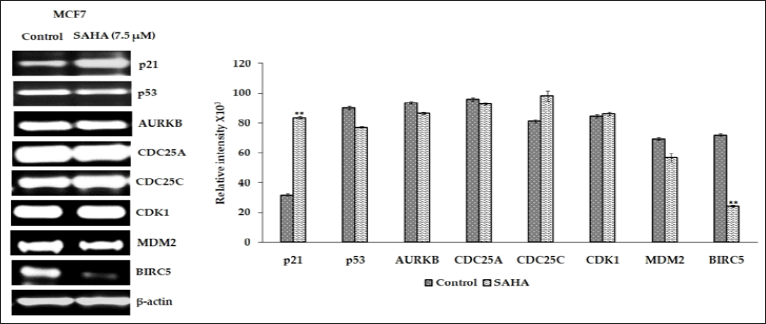
Figure 4: Illustrates that the p21, p53, p-p53, AURKB, CDC25C, CDK1, MDM2 and BIRC5 protein expression in LNCaP-MST cells after SAHA treatment (7.5 μm). Results were analyzed with an unpaired t-test. **: P < 0.01.

Figure 5: Expression levels of p21, p53, p-p53, AURKB, CDC25C, CDK1, MDM2 and BIRC5 protein in MCF-7 cells after SAHA treatments (7.5 μm). The 3-actin level was used as a loading control. Results were statistically analyzed with an unpaired t-test. **: P < 0.01.
The HDAC inhibitors exhibit major anti-proliferative activity against hematological and solid tumors [24,25]. Several antiproliferative effects have been reported for SAHA, including stimulation of G0/G2 cell cycle arrest, differentiation, and selective apoptosis of transformed cells [26-28]. Therefore, the use of HDAC inhibitors as possible therapies for many cancers is gaining momentum as novel research regarding their ability to induce cell cycle arrest and apoptosis are constantly emerges [29,30]. However, the anti-tumor mechanism of HDAC inhibitors are not fully elucidated due to the complex nature of their effects. It has been proven that HDAC inhibitors can selectively affect gene transcription by increasing acetylation of histones and as a result SAHA and Dacinostat (LAQ824), are shown to transcriptionally up- regulate p21 expression. These increases are found to be associated with cell cycle arrest and apoptosis of cancer cells subsequent to HDAC inhibition [31-33]. Both p21 and p27 are multifaceted proteins with functions beyond cell cycle regulation. In addition to regulating the cell cycle, they plays important roles in inducing apoptosis, necrosis, transcriptional regulation, cell migration and cytoskeletal dynamics [34]. Our gene expression experiments in LNCaP-MST and MCF-7 cells demonstrate significant effects on some of the key genes, which play a major role in pathways related to the regulation of cell cycle, both at the mRNA and protein levels.
In the LNCaP-MST cells, the basal level of p21 is expressed at relatively low levels because of MDM2 overexpression. After SAHA treatments, the p21 mRNA and protein levels were increased significantly in LNCaP-MST cells [35]. Similarly, in the MCF-7 cells also p21 protein level was elevated significantly after the SAHA treatment. These elevations, particularly p21WAF1/CIP1 elevation in both LNCaP-MST and MCF-7 cells appears to be p53 independent. Several studies so far have confirmed that SAHA can induce the expression of p21WAF1/CIP1 directly through transcriptional induction. Elevation of p21WAF1/CIP1 has been shown to inhibit the expression as well as the activity of cyclin D1 and causing cell cycle arrest and induction of cancer cell apoptosis [36,37]. Also, the elevation of p21WAF1/CIP1 was found to be coincided with the decreases in AURKB (Aurora kinase B) and Survivin levels due to enhanced acetylation of histone H3 and increased p21WAF1/ CIP1 gene transcription in cancer cells [38]. For example, HDAC inhibitor SAHA was shown to induce p21WAF1/CIP1 expression, and decreased expression of cyclins A, B, and D, as well as their respective Cyclin-Dependent Kinases (CDK), resulting in G1/S and/ or G2/M arrest in cancer cells [39].
During cell cycle regulation p53 is known to play a vital role through impacting multiple pathways. In this regard, the p53 is known to tightly control the expression of p21/WAF1/CIP1 also. However, the p53-independent activation of the p21WAF1/CIP1 promoter by direct SAHA-mediated activation of SP1 sites was clearly evidenced through several reports. Several studies that were focused on p53 independent effects have clearly suggested that SAHA activates the p21 promoter through the Sp1 sites, by stimulating both Sp1 and Sp3 induced gene activation [40]. This activation of p21 is acetylation-dependent mechanism that could account for the often-observed induction of p21 in cells that lack functional p53 [40]. In support of this speculation, our recent study with LNCaP cells was also able to show induction of p21 expression, without any changes in the expression levels of p53, during SAHA induced cell cycle arrest [41]. In consistent with these reports, a significant elevation in the expression of p21WAF1/CIP1 protein was observed in both LNCaP-MST and MCF-7 cells after SAHA treatment, which appears to be not impacted by the p53 status in these cells.
Another mechanism that is commonly reported to cause cell cycle arrest and cell death are mediated through Aurora kinases, a family of serine/threonine kinases, including Aurora A (AURKA), Aurora B (AURKB) and Aurora C (AURKC). These kinases are necessary for cell division since they are involved in the process of chromosomal segregation. AURKB is a protein kinase that ensures the suitable execution and fidelity of mitosis. Enormous evidences have shown that elevation of AURKB expression is involved in cancer progression and in regulating mitosis in various tumors, which makes it as a valuable cancer target that can potentiate the effect of chemotherapeutics [42-44]. In LNCaP-MST cells, AURKB mRNA and protein expressions were significantly down-regulated along with down regulation of CDC25C levels after SAHA treatment. The CDK1 protein expression were down-regulated significantly in LNCaP-MST cells that supported the down-stream mechanism that can be expected from the down-regulation of Aurora Kinase B. Interestingly, the AURKB inhibitions similar to the results observed in LNCaP-MST cells was shown to decrease the expression of Cyclin B1 and Cyclin D1 and elevate the Caspase 3 expression in lymphoma cells leading to cell cycle arrest and apoptosis [45].
In late G1, CDK initiates the expression of G1/S genes by directly phosphorylating and inactivating transcriptional repressors such Rb family members in mammals. Thus, CDK is known to promote the function of transcriptional activators including E2F to stimulate expression of G1/S genes [46-49]. Interestingly, Down-regulation of CDK1 caused by lowered levels of AURKB appears to stop the G1/S transition in LNCaP-MST cells and leading to cell death. On the contrary the AURKB protein levels were found to be elevated in MCF-7 after SAHA treatment, which could be a rebound mechanism in these cells. Overexpression of CDC25A, which is reported in 47% of breast carcinomas, CDC25A was also reported to contribute to the overriding of a physiologic G1 block in breast tumor cells. Thus, the overall results of our study suggest that AURKB and CDC25C axis may contribute to the inhibition of CDK-1 leading to cell cycle arrest and apoptosis in LNCaP-MST cells.
There was a rebound elevation of AURKB and CDC25C in MCF-7 cells. However, there was significant level of cell death after SAHA treatment, which appears to be mediated primarily through p21 in MCF-7 cells. Coincidentally the CDK1 levels were also not altered in MCF-7 cells suggesting that p21 mediate cell death in MCF-7 may not be involving the AURKB - CDC25 axis also.In both LNCaP and MCF-7 cells the expression of survivin was down-regulated after SAHA treatment. According to the previous reports that inhibition of HDAC6 and HDAC3 can be involved to the SAHA induced survivin acetylation, nuclear translocation, and the subsequent protein degradation [50]. Furthermore, inhibition of HDAC3 was shown to precede SAHA induced down-regulation of survivin in treated cells [50]. Thus, surviving may also contribute the cytotoxic effects of SAHA treatment.
Finally, based on our current findings, it can be inferred that the expression patterns of cell cycle-related genes were not similar in LNCaP-MST and MCF-7 cells after SAHA treatment. In LNCaP- MST cells, SAHA treatment down-regulated AURKB, CDC25C and CDK1 coincided with notable elevation of p21 leading to cell cycle arrest and cell death. The current findings in LNCaP-MST cells are in consistence with the previous literature, and therefore it further supports the use of inhibition of HDAC as a significant target for anticancer treatment in prostate cancers with intact p53. However, in MCF-7 cells the changes observed with genes other than p21WAF1/ CIP1 were quite interesting and additional experiments are needed to fully understand the basis for the differences in the intracellular mechanisms.
We would like to thank the Royal Dames of Cancer Research Inc., Ft. Lauderdale, Florida, for their financial support.


