Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Lin Lan* and Xu Xu-Dong
Received: October 18, 2018; Published: October 26, 2018
*Corresponding author: Lin Lan, Department of Bioengineering, School of Medicine, Southeast University, Nanjing, 210009, P. R. China
DOI: 10.26717/BJSTR.2018.10.001964
Production of taxwomen A (TXT), a nitrated dipeptide which inhibits cellulose synthesis in expanding plant tissue, is regarded as the major pathogenicity determinant in Streptomyces scabies, the most widely studied pathogen causing scab disease in economically important crops such as potato. It is known that TXT biosynthetic genes txtA, txtB, txtC and their regulator txtR are clustered together on the pathogenicity island (PAI) of the genome of S. scabies and are conserved in TXT-producing Streptomyces spp. including S. scabies. Here we reported an efficient method to establish the knock-out mutant of txtB (txtBA), by using intergeneric conjugal transfer system constituting Escherichia coli ET12567 /pUZ8002, S. scabies and the vector pKC1139. Briefly, a txtB gene-specific deletion cassette was constructed by PCR and long multiple fusion (LMF), which was subcloned into the Streptomyces-E. coli shuttle vector pKC1139, generating pKC1139-x. By mixing the donor E.coli ET12567 /pUZ8002/ pKC1139-x and the recipient S. scabies at the suitable condition, the exconjugants were successfully obtained and verified by colony PCR. Subsequently, TXT was analyzed for the knock-out mutant (txtBA) in comparison to wild-type of S. scabies. In addition, the transcriptional levels of txtB were studied between the two genotypes. Altogether, txtB gene was successfully knocked out in the exconjugants obtained herein. This would lay a foundation for studying the TXT-biosynthesis associated genes and pathogenicity in scab-causing plant pathogen, facilitating the mechanistic elucidation of plant- pathogen interactions.
Abbreviations: Thaxtomin (TXT); pathogenicity island (PAI); long multiple fusion (LMF); oatmeal broth (OMB); reverse phase(RP); Reverse Transcription (RT); knockout (KO)
Omnipresent in marine and terrestrial habitats, streptomycetes are prolific producers of diverse bioactive metabolites and thus of pharmaceutical and agricultural significance. In spite of a multitude of Streptomyces species characterized to date, only a handful of them are pathogenic to eukaryotic organisms including plants and animals. Among these pathogens, phytopathogens Streptomyces scabies, S. acidiscabies, S. turgidiscabie, draw much research interests because they are known causal agents of common scab diseases in potatoes and other tuber crops such as carrots. The production of phytotoxin thaxtomin A (TXT), an inhibitor of plant cellulose biosynthesis, is regarded to contribute to their pathogenicity [1-3]. TXT has been demonstrated to induce plant cell hypertrophy in expanding tissues. Globally distributed common scab pathogens are represented by S. scabies, the first documented species causing scabs on potatoes [4]. The TXT biosynthetic genes are highly conserved across plant- pathogenic streptomycetes and are found on a pathogenicity island (PAI) that is mobilized among soil-borne streptomycetes, which is considered to be the predominant reason for the emergence of new scab-causing Streptomyces spp [1,5].
To explore and develop the feasible measures to control the rapid spreading of pathogenic streptomycetes in the ecosystem, the molecular mechanism underlying the TXT biosynthesis pathway and the regulation thereof need to be clarified. In this paper we presented an efficient method to establish the knock-out mutant of txtB (txtBA), by using intergeneric conjugal transfer system, consisting of Escherichia coli ET12567 /pUZ8002, S. scabies and the shuttle vector pKC1139. Our study would help deepen the understandings about the mechanism of S. scabies pathogenicity and virulence, thereby providing insight into the elucidation of plant-pathogen interactions.
Streptomyces scabies strain 49173 was grown in the TSB agar (Elite Biotech. Co., Shanghai, China) as previously described [6]. Spore suspensions of Streptomyces strains were prepared as described before [7]. Fresh cultures of Streptomyces cultures grown in TSB tryptone soya broth (tryptone 17g l-1, soya peptone 3g l-1, glucose 2.5g l-1, NaCl 5g l-1, K2HPO4 2.5g l-1) within 48h on a rotary shaker (200rpm, at 30 °C) were used to extract genomic DNA using a bacterial genomic DNA extraction kit (Sangon Biotech Co, Shanghai, China). Streptomyces spores were grown at 26±2 °C in International Streptomyces Project medium 4 (ISP-4) and shaking cultures were harvested for RNA extraction at 4 d. Streptomyces spores were inoculated in oatmeal broth (OMB) [8] for 8-9 d for thaxtomin enrichment and extraction. As for intergeneric conjugation, E. coli strain ET12567 harboring pUZ8002 (E. coli ET12567/pUZ8002) was used as the donor and pKC1139 (Huayueyang Biotech Co., Beijing, China) as conjugative vector. E. coli ET12567/pUZ8002 and E. coli 12567/pKC1139 were maintained in LB medium (Luria- Bertani broth, Sangon Biotech Co, Shanghai, China) supplemented with apramycin (Apra, 50μg ml-1), kanamycin (Kana, 50μg ml-1) and chloramphenicol (Cm, 25μg ml-1).
The genomic DNA was prepared from 48h shaking culture of S. scabies by using EZ-10 spin column bacterial genomic DNA isolation kit (Sangon Biotech Co, Shanghai, China), with the addition of lysozyme solution (20mg ml) which facilitated the removal of Streptomyces cell wall.
Construction Of Plasmids For Gene Deletion: A txtB deletion mutant was created using PCR-based homologous recombination method [9] by replacing the gene of interest with a txtB gene-specific deletion cassette. A thiostrepton resistance gene [tsr (ThioR)] from the plasmid pIJ8600 was PCR-amplified using the primers P3 and P4 (Table 1), resulting in a 1058bp fragment. A 428bp fragment designated txtB_1, encompassing the nucleotides upstream of txtB gene CDS (coding sequence) and some of its CDS, was PCR-amplified from the S. scabies genomic DNA using the primers P1 and P2 (Table 1). A 531bp fragment designated txtB_2, encompassing the some of txtB gene CDS and the nucleotides downstream of this gene, was amplified from the S. scabies genomic DNA using the primers P5 and P6 (Table 1). The three fragments were ligated according to the LMF method described by Shevchuk et al. [10], yielding fragment txtB1-tsr-txtB2. A deletion cassette containing the upstream homologous arm (~420bp), and the downstream homologous arm (~530bp), between which is the tsr marker gene (ThioR) was PCR-amplified with the txtB gene-specific primer pairs that carry BamHI, HindIII site sequences, respectively using the fragment txtB1-tsr-txtB2 as the template. The obtained deletion cassette was cloned into pMD19T Simple (Takara Biotech Co., Dalian, China), followed by BamHI and HindIII digestion, ligation into BamHI Hindlll digested linearized pKC1139, generating the txtB deletion vector pKC1139-x.
Intergeneric Conjugation and Identification of Exconju- gants: The donor cells of E. coli ET12567 /pUZ8002 containing pKC1139-1 were grown to an OD600 of 0.4-0.6 with the appropriate antibiotics. MS medium (mannitol 20g L-1, soya flour 20g L-1) containing 10mM MgCl2 and TSB medium were used for plating of E. coli/Streptomyces scabies in intergeneric conjugation and the efficiency of conjugation was evaluated between MS and TSB. The conjugation plates were incubated at 28 °C for 16-20h, and subsequently overlaid with 1.5mL of sterile water containing 500μg mL-1 nalidixic acid and 1000μg mL-1 thiostrepton, which were incubated further for 10-12d, until the exconjugants emerged. ApraR (100μg mL-1) exconjugants were screened for CmS (25μg mL-1), indicating a double-crossover allelic exchange in S. scabies, and were subsequently verified by colony PCR.
TXT Extraction and Assay: S. scabies cultures of wildtype and mutant were grown for 8 d and cells were pelleted by centrifugation (10000xg). The supernatants were extracted with ethyl acetate, followed by evaporation, and the residues were dissolved in methanol, and analyzed for the presence of TXT using an Agilent 1200 HPLC (Agilent Technologies, CA, USA) on a C-18 reversed-phase(RP) column eluting with a gradient series of 20% to 100% methanol in water as the mobile phase.
Transcriptional Analysis of txtB With RT-PCR: RNA was extracted from 3d mycelia grown on ISP-4 at 28 °C using EZ-10 total RNA Mini-Preps Kit (Bio Basic Canada Inc. Ontario, Canada). Genomic DNA was removed from total RNA using DNAse I (Bio Basic Canada Inc. Ontario, Canada). DNAse-treated RNA (1μg) was reverse transcribed using the Superscript™ III 1st stand synthesis system kit and random hexamer primers (Invitrogen, USA) according to the manufacturer's protocol. Gene expression was verified by PCR amplifying the cDNA using gene-specific primers. The gyrA gene (NZ_LRTN01000850.1:272-2866, http://www.sanger.ac.uk/ Projects/S_scabies/), which encodes DNA gyrase subunit A, was used to normalize the amount of RNA in the samples. Each measurement was performed in triplicate.
We aimed to construct a pKC1139-based vector, designated pKC1139-x, which carried gene-specific homologous upstream and downstream arms for knock-out purpose (Figure 1). By PCR amplification, the txtB gene-specific upstream and downstream fragments [homologous arms, (Figure 2a)] and the tsr, a thiostrepton resistance marker (Figure 2b), were obtained respectively. The individual band was purified using DNA gel extraction kit for further use.
Figure 1: Schematic diagram showing the construct of gene (txtB) inactivation mutants via the homologous recombination.
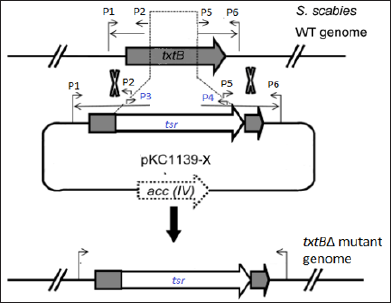
Figure 2: Electrophoretic gel analysis.
Note:
a) PCR products of txtB_1, and txtB_2, sized 428bp and 531bp, respectively.
b) PCR amplification of tsr gene, a thiostrepton resistance marker, using pIJ8600 as the template. M= D2000 DNA ladder (Tiangen Biotech. Co., Bejing, China)
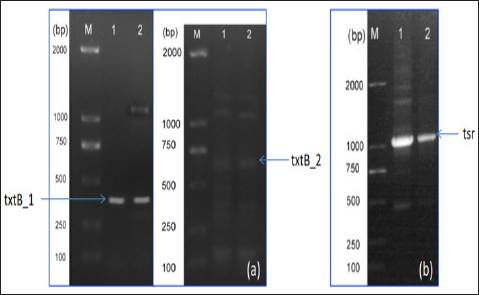
By adopting the long multiple fusion (LMF) method as described in Shevchuk et al. [10], the three fragments (txtB_1, tsr, txtB_2) were ligated together to generate a long fragment (~2kb) (Figure 3). This long fragment (sized ~2kb) was used for gene inactivation in S. scabies based on homologous recombination.
As demonstrated by previous studies, OMB was the suitable medium for S. scabies to biosynthesize TXT [8]. Following culturing in OMB for 8-9d, the culture supernatants of S. scabies txtBA mutant and WT were subjected to organic-solvent extraction. Subsequently, TXT levels of both genotypes were determined against TXT standard by reverse phase(RP)-HPLC. As seen from the chromatographs, no TXT was detected in txtBA (Figure 4a), suggesting that the production of TXT was abrogated for txtBA as compared to WT which produced normal level of TXT (Figures 4b & 4c).
Figure 3: Electrophoretic gel of the fused long fragment (sized 2017bp) using LMF method. M: D15000 DNA ladder (Tiangen Biotech. Co., Bejing, China). Lane 1 and 2 indicates the sample 1 and 2, respectively.
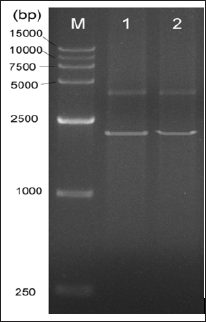
Figure 4: Reverse-phase(RP)-HPLC chromatographs of TXT extracted from OMB broth.
Note: a) txtBΔ mutant;
b) WT of S. scabies, and
c) TXT standard.
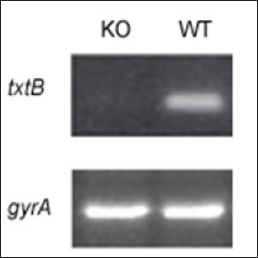
In view of that txtB deletion generates txtB knockout (KO) mutant, it seemed reasonable to question whether KO abolished transcription of txtB. In this study, the transcriptional level of txtB was monitored by reverse transcription (RT) -PCR with the txtB gene-specific primers. As shown in Figure 5, transcription of txtB was diminished in KO mutant while WT displayed abundant transcripts of txtB. In summary, an efficient intergeneric DNA transfer system was successfully developed between E. coli and Streptomyces sp. This method would lay a basis for DNA transformation and mutational studies of economically important strain S. scabies and other Streptomyces species as well.
Figure 5: Relative abundance of txtB transcripts as assayed by RT-PCR from S. scabies WT versus txtBA (knockout, KO) mutant. The gyrA gene, was used as an internal control to normalize the amount of cDNA template in RT- PCR analysis.
This work was supported by a funding from the Natural Science Foundation of Jiangsu Province, China (Grant No. BK20151133) to Lin L.


