Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Laura Berbegal*1 , Juan Carlos Rodriguez2, Jose Sanchez Payá3 , Antonio Jose Galiana4 , Francisco Jose De Leon5 , Maria Paz Ventero2, María Teresa Martínez Miravete6 and Isabel Betlloch7
Received: October 30, 2018; Published: November 08, 2018
*Corresponding author: Laura Berbegal De Gracia, Hospital Marina Salud de Dénia, Av Marina Alta s/n, 03700 Dénia (Alicante), Spain
DOI: 10.26717/BJSTR.2018.10.002014
The arise of next generation sequencing (NGS) has improved the knowledge about the cutaneous and nasal microbiota in the development of atopic dermatitis (AD). Recently, it has been demonstrated that the microbiota plays a key role in this pathology. For our study we compared the nasal microbiome in children with AD versus healthy controls. Many different bacteria were found in this ecosystem; Staphylococcus aureus would appear to play a lesser role than suggested in studies based on nasopharyngeal cultures. However, the genus Oscillospira was more common in the children with AD and was also associated with lower body mass index. Further research is needed to understand the clinical importance of these findings in improving disease management
Keywords: NGS; Atopic Dermatitis; Microbiota; Oscillospira Spp; Obesity
Until a few years ago, little was known about the relationship between the skin microbiota, skin health and immune response, as research in this area had only focused on the microorganisms classically known to be pathogens [1]. The recent development of molecular microbiology techniques, especially next generation sequencing (NGS), has revolutionized this area [2], and more and more studies are shedding light on the interactions between the host and the microbiota, and their relationship with atopic dermatitis (AD) [3-5]. Today, it is known that the maturation of the microbiota in the nostrils is relevant in the evolution of AD, a condition whose incidence and severity changes depending on a person’s physiological maturity and improves notably following puberty and in adulthood. At the same time, in adolescents and adults, Staphylococcus and Streptococcus in the nostrils tend to be replaced by lipophilic bacteria, which could explain the decreased incidence of AD and nasal colonization with S. aureus with age [6,7].
The skin microbiota can also change during AD flare-ups, depending on variables like recent treatments and the host’s inflammatory response, among others. Kong and colleagues found an association between AD flare-ups and a decrease in the diversity of the skin microbiota [8]. In fact, they observed that the proportion of Staphylococcus spp. increases and microbiota diversity decreases prior to an exacerbation of AD [8]. On the other hand, active treatment with anti-inflammatories helps to control the disease, favoring the restoration of microbiota diversity (especially the population of Streptococcus spp., Propionibacterium spp. and Corynebacterium spp.) and the limited levels of Staphylococcus in the post-flare-up period following treatment [9]. One study even found that treatment with emollients can reduce the proportion of S. aureus and increase microbiota diversity [10]. This study aimed to describe the composition, abundance and dominance of species; bacterial communities; and the biodiversity of the nasopharyngeal microbiota in children with and without AD.
In this study, the nasopharyngeal microbiota in 8 children with AD (cases) and in 10 without (controls) was analyzed. Inclusion criteria for cases were: aged 6 months to 16 years, attending the Pediatric Dermatology Unit of the General University Hospital of Alicante (HGUA in its Spanish abbreviation), and diagnosis of AD based on the consensus criteria of the American Academy of Dermatology [11]. Controls were children of the same age who also attended our unit during the same period but did not present with AD or any other infectious or dermatological disease that could induce diagnostic confusion, such as contact dermatitis, scabies, or frictional infantile dermatitis. For this study, participants with another systemic disease, antibiotic or immunosuppressant treatment in the previous 30 days, or a recent history of infections like erysipelas or cellulitis were excluded.
After obtaining informed consent from the participant’s parent or legal guardian for study inclusion, the study variables were recorded during the pediatric dermatology consultation using a purpose-designed data extraction form (patient identifier; case/ control; age; sex; height; weight; body mass index [BMI]; family history of AD/asthma/rhinitis; personal history of rhinitis/ asthma/inhaled allergies/food allergies/celiac disease; SCORAD; and, in cases, sleep disturbances due to pruritus). Data were coded, anonymized and stored in a database using SPSS software (version 22). The HGUA ethics committee approved the study. Initially, 12 cases and 12 controls satisfied the inclusion criteria proposed in this study, but only the data of 8 cases and 10 controls were finally included, because the other nasal samples were insufficient for processing and analysis. For the microbiological study, samples were collected using nasopharyngeal aspirate, by means of the vacuum-assisted method. Specimens were analyzed using NGS technique to detect the presence of 16S rRNA. To this end, bacterial DNA was extracted and purified using the Molzym MolYsis™ system. To enrich the quantity of existing bacterial DNA in each sample.
Then, a 16S ribosomal gene amplification by PCR was carried out for each nasal sample following the protocol “Metagenomic 16S” described by Illumina [12]. Sequencing was performed by means of the Illumina MiSeq system, using 300x2 V3 reagents (pairend). Once the sequences were obtained, they were analyzed using the SSU-ALIGN and Meta-RNA programs. Finally, the sequences were compared with the GreenGenes [13] y RDP databases [14], using BLAST. The comparison of the samples against each other was undertaken using the Unifrac Software, which was specially designed by the International Human Microbiome Consortium for analyzing ribosomal genes of microorganisms from human tissue. Using Qiime Software (version 1.8.0), the bioinformatics study on the samples were carried out to quantify the absolute and relative abundance of species, determine the dominance and composition of the bacterial communities, perform a study of the biodiversity, and analyze the principal components identified.
The abundance analysis of individual genera in cases and controls (Figures 1a-1d) shown that the bacterial communities in the nasopharyngeal aspirate samples contained many different genera and were not dominated by Staphylococcus (Figure 2). Nor was Staphylococcus significantly more abundant in the cases than in the controls (Figure 3a). Also, different relative abundance of species between controls and cases were found. Highlight, Oscillospira spp. were significantly more abundant in the children with AD (1.7%) than in those without this pathology (0.2%) (Pvalor = 0,03) (Figures 1e & 3a). Moreover, the cases, which had more relative abundance of Oscillospira spp., shown a lower BMI than the controls on average. The descriptive analysis of the BMI data revealed that the 50% of the controls and only the 25% of the cases were overweight or obese. This data could suggest that Oscillospira spp presence is associated to a lower risk of obesity. The four dominant phyla in the individual samples taken from our cases and controls were Firmicutes, Proteobacteria, Bacteroidetes and Actinobacteria (Figures 2a-2d). The Chao1 and Shannon diversity index shown greater diversity in the samples, taken from the cases than in those taken from the controls (51 vs 34 and 3.29 vs 1.14 respectively), though these differences do not reach statistical significance (p= 0.23 and p= 0.37, respectively) (Figure 3b). None of the variables included in our study explain why a cluster of the participants had a similar microbiome (Figure 3c).
Figure 1: Bioinformatics analysis of genera.
Note:
a. Segmented bar graph showing the relative abundance of the different genera in each case.
b. Table showing the individual relative abundances of the different genera in each case.
c. Segmented bar graph showing the relative abundance of the different genera in each control.
d. Table showing the individual relative abundances of the different genera in each control.
e. Table comparing the individual relative abundances of the different genera in cases and controls.
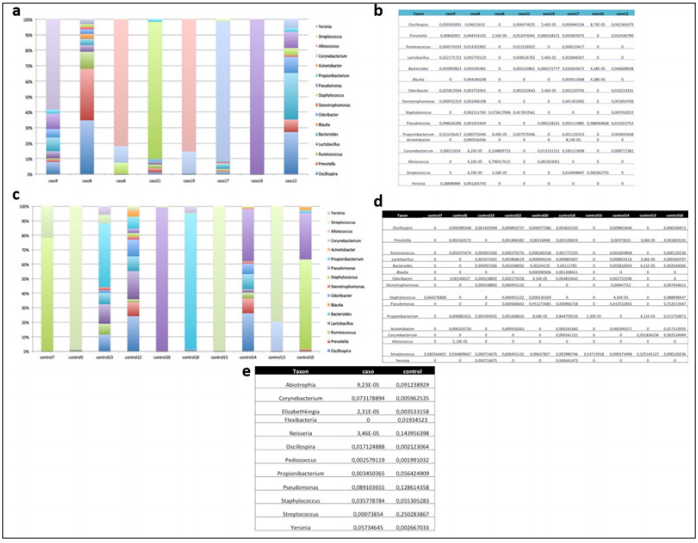
Figure 2: Bioinformatics analysis of phyla.
Note:
a. Segmented bar graph showing the relative abundance of the different phyla in each case.
b. Table showing the individual relative abundances of the different phyla in each case.
c. Segmented bar graph showing the relative abundance of the different phyla in each control.
d. Table showing the individual relative abundances of the different phyla in each control.
e. Segmented bar graph comparing the abundance of phyla in cases and controls.

Figure 3: ANOVA analysis, diversity indices and principal component analysis.
Note:
a. Table showing ANOVA analysis of the genera with statistical significance between cases and controls, and of the genus Staphylococcus.
b. Chao1 and Shannon diversity indices, comparing cases and controls; Figure 3c: Principal component analysis (cluster of participants with similar microbiota).

Oscillospira is a bacterium that has never been cultured and whose role in AD is unknown, as there have been no studies to date investigating its involvement in this pathology, although there have been studies of its interaction with the immune system [15,16]. Our study is the first to suggest that the presence of this microorganism in the nasopharyngeal tract is associated with AD, and it stands in contrast to other study that have described its lower presence in the gut of patients with food allergy [17]. On the other hand, there are many studies that link the presence of Oscillospira spp. in the intestines with a decreased risk of obesity [18-20]. Our results are in line with this affirmation, as the cases had lower mean BMI than the controls, and half of the controls (5/10) were overweight or obese, compared to a quarter (2/8) of the cases. The presence of this microorganism at nasopharyngeal level in patients with AD presents new challenges for research into this disease, and new opportunities for alternative treatments targeting the microbiota. However, more studies are necessary to definitively establish the role of Oscillospira in AD.


