Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Emmanuel G Pintelas1 and Panagiotis Pintelas*2
Received: November 05, 2018; Published: November 19, 2018
*Corresponding author: Panagiotis Pintelas, Department of Mathematics, University of Patras, Greece
DOI: 10.26717/BJSTR.2018.11.002066
In recent years machine learning has been thoroughly used in the bioinformatics and biomedical field. The prediction of cellular localization of the proteins can be considered very significant task in bioinformatics since wrong localization site can cause various diseases and infections to humans. Ensemble learning algorithms and semi-supervised algorithms have been independently developed to build efficient and robust classification models. In this paper we focus on the prediction of protein localization site in Escherichia Coli and Saccharomyces cerevisiae organisms utilizing a semi-supervised self-labeled algorithm based on ensemble methodologies. The experimental results showed the efficiency of our proposed algorithm compared against state-of-the-art self-labeled techniques.
Proteins are important molecules in our cells made up of long sequences of amino acid residues [1]. Each protein within the body has a specific function, while they work normally when they are in the correct localization site. The function of a protein in general can be affected by its cellular localization (the location a protein has in a cell) and contributes to many diseases like cardiovascular, metabolic, neurodegenerative diseases and cancer [2]. Also, it is of high interest in various research areas, like therapeutic target discovery, drug design and biological research [3]. Therefore, the prediction of cellular localization of the proteins can be considered very helpful and is a significant task in bioinformatics which has been studied a lot [4-6].
In general, a prediction tool can take as input some attributes of a protein such as its protein sequence of amino acids and predict the location where this protein resides in a cell, such as the nucleus and Endoplasmic reticulum. X-ray crystallography, electron crystallography and nuclear magnetic resonance are some traditionally biochemical experimental methods adopted [7] for predicting protein cellular location. These methods are accurate and precise in general, but they are inefficient and unpractical because they are expensive and time consuming. Therefore, in the last two decades computational methods especially using machine learning methods have been developed to make predictions [5,8-17]. Escherichia Coli (E. coli) and Saccharomyces cerevisiae (Yeast) are two well characterized unicellular organisms which have been exhaustively studied [18]. These two organisms have different proteins allocated in their cell where they must be at their accurate positions. A wrong localization site of these proteins in the cell can cause various diseases and infections to humans such as bloody diarrhea [19].
In the past, there have been significant efforts for predicting the localization sites of proteins [18-28]. Anastasiadis and Magoulas [18] investigated the performance of K nearest neighbours, feed-forward neural networks with and without cross-validation and ensemble-based techniques for the prediction of protein localization sites in E. coli and Yeast. Their results showed that the ensemble-based techniques had the highest average classification accuracy per class, achieving 91.7% and 66.2% for E. coli and Yeast respectively. Chen [22], implemented three different machine learning techniques: Decision tree, perceptrons, two-layer feed-forward neural network for predicting proteins’ cellular localization on E. coli and Yeast datasets. From the results, a similar prediction accuracy was found for all three techniques and 65%~70% on E. coli dataset and 46%~50% on Yeast dataset. Sengur [23], investigated the performance of an artificial immune system based on fuzzy k-NN algorithm. The highest average classification accuracy was 97.29% for E. coli and 76.4% for Yeast. Bouziane et al. [21], utilized four supervised machine learning algorithms for the prediction of cellular localization sites of proteins. For their experiments, they used Naïve Bayesian, k-Nearest Neighbour and feed-forward neural network classifiers.
The highest classification accuracy they managed to achieve was 95.8% for E. coli dataset and 73.4% for Yeast dataset. Very recently Priya and Chhabra [19], proposed a hybrid model of Support Vector Machine and the LogitBoost technique for the prediction of the protein localization site in E. coli bacteria. The maximum classification accuracy achieved was 95.23%. Motivated by previous work Satu et al. [20], utilized E. coli and Yeast datasets for the problem of protein localization prediction. For their experiments they used several data mining classification algorithms which were: lazy classifiers (kNN, KStar), meta classifiers (Iterative Classifies Optimizer, Logit boost, Random Committee, Rotation Forest), function classifiers (Logistics, Simple Logistics), tree classifier (LMT, Random Forest, Random Tree) and artificial neural networks, achieving 87.50% with Rotation Forest and 60.53% with Random Forest maximum classification accuracy for E. coli and Yeast respectively.
Nevertheless, the problem of prediction of protein localization sites is considered a challenging task since finding labeled data is often an expensive and time-consuming procedure [29], as it requires human efforts. To address this problem, Semi-Supervised Learning (SSL) algorithms utilize both labeled and unlabeled data since in general finding sufficient unlabeled data is significantly easier than finding labeled data [30-32]. The basic aim of SSL is to exploit the hidden information found in the unlabeled data in order to train classifiers more efficiently [33,34]. The most popular SSL algorithms are self-labeled algorithms. These algorithms make predictions on a large amount of unlabeled data aiming to enlarge a small amount of labeled data. Triguero et al. [35] made a taxonomy of self-labeled algorithms based on their main characteristics and conducted a comprehensive research of their classification efficacy on several datasets. Some of the most efficient and popular Selflabeled algorithms proposed in the literature are Self-training [30], Co-training [31], Tri-training [35], Democratic-Co learning [37], Co- Forest [38] and Co-Bagging [39].
In Self-training, one classifier following an iterative procedure is trained on a labeled dataset which is augmented by its most confident predictions on an unlabeled dataset. In Co-training, two classifiers are trained separately using two different views on a labeled dataset and then each classifier adds the most confident predictions on an unlabeled dataset to the training set of the other. Tri-training algorithm utilizes three classifiers which teach each other based on a majority voting strategy. Democratic-Co learning utilizes several classifiers following a majority voting and confidence measurement strategy for predicting the values of unlabeled examples. Co-Forest algorithm trains Random trees on bootstrap data from the dataset assigning few unlabeled examples to each tree, utilizing a majority voting. Co-Bagging algorithm trains multiple base classifiers on bootstrap data created by random resampling with replacement from the training set.
Ensemble Learning (EL) is a different approach, which has been developed in the last decades, for building more efficient composite global model by the combination of several prediction models than using a single one [40]. Moreover, the combination of SSL and EL are beneficial to each other [41], leading to even better classification results by developing more accurate and robust classifiers [42-47] than utilizing EL and SSL independently. Recently, Livieris et al. [43,45] proposed some ensemble SSL algorithms which utilize the individual predictions of the most popular self-labeled methods i.e. Self-training, Co-training and Tri-training based on a combination of various voting strategies. Motivated by previous work, Livieris et al. [48] proposed a new semi-supervised learning algorithm which selects the most promising base learner from a number of classifiers utilizing a Self-training methodology.
In this work, we propose a semi-supervised self-labeled algorithm based on the ensemble approach for the prediction of protein localization sites on E. coli and Yeast organisms. The proposed algorithm constitutes a modification of the CST-Voting, utilizing each self-labeled algorithm with the base learner, which presents the highest accuracy. It is worth mentioning that we utilized only a 10%-50% ratio of the training set in our experiments in order to evaluate the efficiency of the SSL approach. Our experimental results reveal the efficiency of the proposed algorithm compared against state-of-the-art self-labeled methods. The remainder of this paper is organized as follows: Section 2 presents the proposed classification algorithm and a brief description of the data utilized in our study. Section 3 presents a series of experiments in order to evaluate the accuracy of the proposed algorithm against the most popular self-labeled classification algorithms. Finally, in Section 4 we present our concluding remarks.
The main goal of the research described in this paper is the development of a prediction model for the classification of protein localization site in Escherichia Coli (E. coli) and Saccharomyces Cerevisiae (Yeast) organisms utilizing a semi-supervised selflabeled algorithm. For this purpose, we adopted a two-stages methodology, where the first stage deploys the self-labeled classification algorithm while the second one concerns dataset utilized in this study.
In this section, we present a detailed description of the proposed SSL algorithm for the prediction of protein localization, which is based on an ensemble philosophy, entitled CST*-Voting. Recently, Livieris et al. [43], proposed the CST-Voting algorithm which combines the self-labeled framework along with ensemble learning techniques. In particular, this algorithm exploits the individual predictions of the most popular self-labeled algorithms namely, Co-training, Self-training and Tri-training utilizing simple majority voting. These self-labeled methods operate in a different way to take advantage of the hidden information found in the unlabeled data in order to enlarge a labeled dataset. The main difference between these self-labeled algorithms is the technique used to exploit the unlabeled data. More specifically, self-training and tri-training are single-view methods, while co-training is a multi-view method. Furthermore, it is worth mentioning that co training and tri-training are indeed ensemble methods, since they both make use of multiple classifiers.
Along with this line, we consider to improve the classification efficiency of the ensemble, by utilizing each self-labeled algorithm with the base learner, which presents the highest accuracy. To this end, Co-training utilizes Sequential Minimum Optimization (SMO) [49] as base learner, Self-training utilizes Multilayer perceptron (MLP) [50] and Tri-training utilizes C4.5 [51]. The motivation for this selection is based upon the fact that these algorithms were reported to present the best efficiency using these specific base learners [35,43]. A high-level description of the proposed CST*-Voting is presented in Algorithm 1. Initially, the classical semi-supervised algorithms, which constitute the ensemble, i.e., self-training (MLP), co-training (SMO) and tri-training (C4.5), are trained utilizing the same labeled and unlabeled U dataset. Subsequently, the final hypothesis on an unlabeled example of the test set combines the individual predictions of the self-labeled algorithms, hence utilizing a majority voting. Clearly, the ensemble output is the one made by more than half of them. An overview of proposed algorithm is depicted in Figure 1.
Algorithm 1. CST*-Voting
Input: L- Set of labeled instances.
U- Set of unlabeled instances.
T- Training set.
Output: Trained ensemble classifier
(Phase I: Training)
1: Self-training(L, U) using MLP as base-learner.
2: Co-training(L, U) using SMO as base-learner.
3: Tri-training(L, U) using C4.5 as base-learner.
(Phase II: Voting)
Comment: The labels of instances in the testing set are placed in L through steps 4 - 7.
4: for each x ∈ T do
5: Apply the trained classifiers on x.
6: Use majority vote to predict the label y* of x.
7: end for
In the experiments were used E. coli and Yeast datasets for the localization of proteins. Both are collected from UCI Machine learning data repository. E. coli dataset has 336 instances which are labeled into 8 classes. The attributes on this dataset are mcg, gvh, lip, chg, aac, alm1, alm2 which all are numeric. There are eight classes namely: CP (Cytoplasm), IM (Inner Membrane without signal sequence), PP(Periplasm), IMU (Inner Membrane with Uncleavable signal sequence), OM (Outer Membrane), OML (Outer Membrane Lipoprotein), IML (Inner Membrane Lipoprotein), IMS (Inner Membrane with cleavable signal sequence). Yeast dataset has 1462 instances which are labeled into 10 classes. The attributes on this dataset are mcg, gvh, alm, mit, erl, pox, vac, nuc which all are numeric. There are ten classes namely: Cytoplasmic (CYT), Nuclear (NUC), Vacuolar (VAC), Mitochondrial (MIT), Peroxisomal (POX), Extracellular (EXC), Endoplasmic Reticulum (ERL), Membrane proteins with a cleaved signal (MEl) membrane proteins with an uncleaved signal (ME2) and membrane proteins with no N-terrninal signM (ME3).
Next, we focus our interest on the experimental analysis for evaluating the classification performance of CST*-Voting against the most efficient and frequently utilized self-labeled methods, i.e. Self-training, Co-training, Tri-training. Notice that all selflabeled methods deployed base learners the SMO, the C4.5 and the MLP algorithm. These supervised classifiers probably constitute the most effective and popular machine learning algorithms for classification problems [50]. All self-labeled algorithms utilized the configuration parameter settings as in [44-48] and all base learners were used with their default parameter settings included in the WEKA 3.9 library [51] in order to minimize the effect of any expert bias, instead of attempting to tune any of the algorithms to the specific dataset. Furthermore, in order to study the influence of the amount of labeled data, five different ratios (R) of the training data were used, i.e. 10%, 20%, 30%, 40% and 50%. Tables 1 & 2 present the performance of all self-labeled methods on E. coli dataset and Yeast dataset, respectively. Notice that the highest classification performance for each labeled ratio and performance metric is highlighted in bold.
The aggregated results showed that the CST*-Voting was by far the most efficient and robust method independent of the utilized ratio of labeled instances in the training set, performing better in all five ratio cases for both datasets. Furthermore, a more representative visualization of the classification accuracy of all compared SSL algorithms is presented in Figures 2-7 for E. coli and Yeast datasets respectively. In more detail, these figures present a performance comparison of CST*-Voting versus Self-training, Co-training, Tri-training, CST-Voting which utilize C4.5, SMO and MLP as base learners. Each box-plot presents the accuracy measure for each tested SSL algorithm according to the labeled ratio.
Figure 2:Performance evaluation of each SSL algorithm using C4.5 as base learner versus CST*-Voting for E. coli dataset.
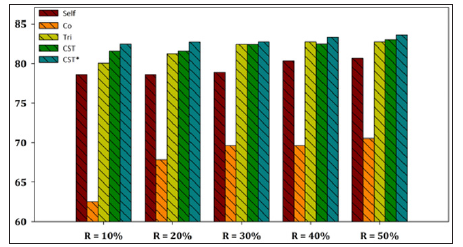
Figure 3: Performance evaluation of each SSL algorithm using MLP as base learner versus CST*-Voting for E. coli dataset.
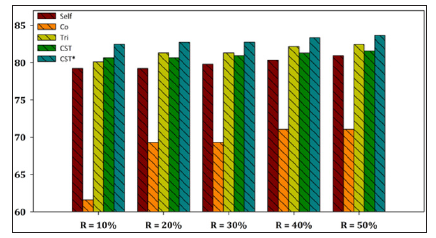
Figure 4: Performance evaluation of each SSL algorithm using SMO as base learner versus CST*-Voting for E. coli dataset.
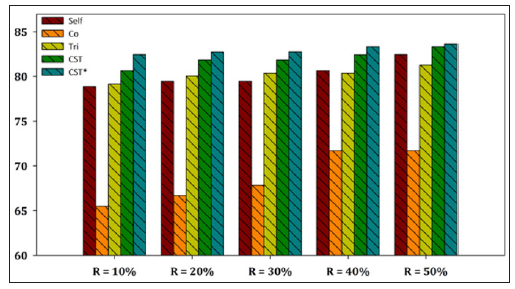
Figure 5: Performance evaluation of each SSL algorithm using C4.5 as base learner versus CST*-Voting for Yeast dataset.
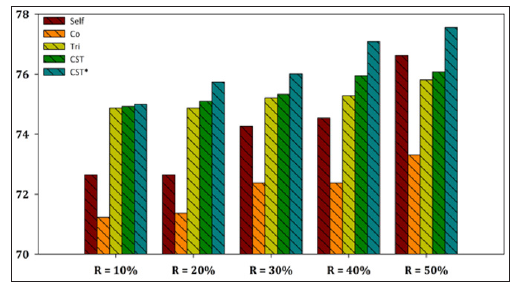
Figure 6: Performance evaluation of each SSL algorithm using MLP as base learner versus CST*-Voting for Yeast dataset.
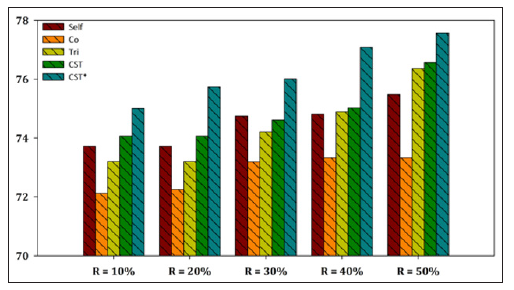
Figure 7:Performance evaluation of each SSL algorithm using SMO as base learner versus CST*-Voting for Yeast dataset.
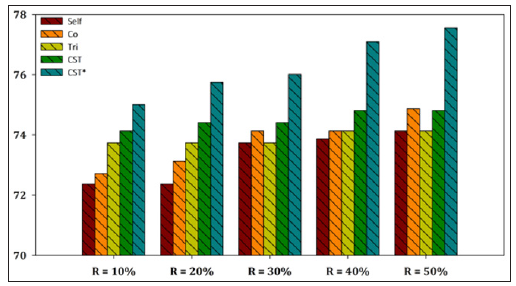
In this work, we evaluated the performance of an ensemblebased self-labeled algorithm for protein localization sites, called CST*-Voting using two datasets (E. coli and Yeast). The proposed algorithm constitutes a modification of the CST-Voting, utilizing three self-labeled algorithms i.e. Self-training, Tri-training and Co-training, using the base learner which presented the highest accuracy in literature. A series of experiments were carried out in order to evaluate the classification performance of the proposed algorithm against the most efficient and frequently utilized selflabeled methods. To this end, we utilized only a 10%-50% ratios of the training set in our experiments, instead of the entire dataset, in order to evaluate the efficiency of the SSL approach. As our experimental results have shown, the efficiency of the proposed algorithm is better compared against state-of-the-art self-labeled methods. In our future work we intend to invest on extending our experiments of the proposed algorithm to several organism’s cells for protein localization prediction and on improving the prediction accuracy of ensemble SSL utilizing more efficient and sophisticated self-labeled algorithms.


