Abstract
The effects of Low-Dose Radiation (LDR) on humans and other organisms are still debated, even though LDR is ubiquitous in our environment. One prominent phenomenon of LDR is Hyper-Radiosensitivity (HRS), which may increase cancer risk. We reported previously that the radiation dose conferring HRS was 75 mGy with γ-rays from a 60Co source in human normal fibroblast cells. To examine whether LDR causes any alterations in genomic DNA, especially in cancer-related genes, high-throughput next generation sequencing (NGS) was adopted. Whole genome sequencing (WGS) of the genomic DNA from non-irradiated control cells, 75 mGy irradiated cells, and 10,000 mGy irradiated cells, identified alterations in several genes specific to LDR-irradiated cells. Amplicon sequencing did not identify apparent alterations in any hotspots of 50 cancer-related genes.
Keywords: Low-Dose Radiation; Ionizing Radiation; Human Fibroblast Cells; Genomic Alteration; Cancer-Related Gene; Next Generation Sequencing
Introduction
The effects of Low-Dose Radiation (LDR) on human health are still debated, despite LDR being ubiquitous in our environment, such as via radioactive substance leakage following an accident at a nuclear facility, as well as medical exposure for diagnosis and radiotherapy. Recent research has suggested that the biological effects of LDR differ from those observed by High-Dose Radiation (HDR) [1]. Both LDR and HDR could have the potential to initiate cancer development in organisms because most cancers are caused by the initial alterations of oncogenes, anti-cancer genes, DNA repair-relevant genes, cell attachment genes, and cell migration genes, etc. We reported in a previous paper that the dose conferring HRS was 75 mGy with γ-rays from a 60Co source in human normal fibroblast cells [2]. In recent years, research concerning the effects of LDR and HDR on the genome has been increasing and the research can be applied for the diagnosis and cure of cancers [3-8]. WGS technologies make it possible to analyze the entire genome shallowly and amplicon sequencing makes it possible to read limited genes deeply. However, research utilizing the widely used human normal fibroblast cells to detect the effect of LDR on DNA is scarce [9]. In this study, high-throughput NGS was used to search for alterations in the genomic DNA, especially cancer-related genes and genetic disease-related genes, caused by LDR. Using nonirradiated control, 75 mGy (LDR) irradiated, and 10,000 mGy (HDR) irradiated human normal fibroblast cells, WGS was performed to scan all identified genes. Next, cancer-panel amplicon sequencing was performed to scan cancer-related genes whose hot spots are known in many types of cancers.
Materials and Methods
Cell Culture and Irradiation by Γ-Rays
The Normal Human Dermal Fibroblasts (NHDF) came from an adult donor and were purchased from KURABO (Osaka, Japan). After thawing, NHDFs were routinely cultivated in T-25 flasks (BD Biosciences, Bedford, MA, USA) with phenol red-free Dulbecco’s modified Eagle’s medium (D2902, Sigma-Aldrich, St. Louis, USA) supplemented with 10% glucose for a final concentration of 4.5 g/L, 5% sodium carbonate to 3.7 g/L, fetal bovine serum (Invitrogen Corp., CA, USA) to 10%, penicillin (Wako Pure Chemical Industries) to 100/U, and streptomycin to 100 μg/mL (Wako Pure Chemical Industries). The cells were cultivated at 37OC in an atmosphere of 95% room air and 5% carbon dioxide. When the cells were grown to 80% confluence, they were detached by trypsin, counted, and cultivated in 3.5-cm dishes with a density of approximately 1.0-2.0 × 104 cells/dish. After 2 days from seeding, cells were irradiated with gamma rays from a 60Co source at the Radiation Research Center of Osaka Prefecture University, Japan.
The irradiation dose was defined by the distance of each dish from the 60Co source, and cells were irradiated with doses of 75, and 10,000 mGy with dose rates of 1.5 and 200 Gy/h, respectively, at room temperature. The 0 mGy (control) group was transported to the staff-waiting room to discriminate observations from environmental stress.
Next Generation Sequencing
After 2 days of irradiation and cultivation, genomic DNA was purified using Nucleospin Tissue (#740-952-50 CloneTech/ Takara, Kusatsu, Japan). Analysis by WGS was trusted to Filgen, Inc. (Nagoya, Japan) as the Variation Detection Project WGS. Analysis by amplicon sequencing was trusted to Takara Bio Inc. (Kusatsu, Japan) as the Ion AmpliSeq Cancer Panel. QC control of the three DNA samples were passed for further analysis. Information of the genes were referenced by PubMed or GeneCards.
Data Mining
Data from the three groups (control [0 mGy], LDR [75 mGy] and HDR [1,000 mGy]) were compared using Excel included in Office 2010 (Microsoft Inc., USA). We concentrated on the alterations specific to LDR, specific to HDR, or common to LDR and HDR but not found in the control group. At first, for WGS, alterations among the exons and those cause frame-shifts from the reference human genome sequence, hg19 [10] were selected, because the alterations of in-frame usually cause no or scarce effect. Among genomic alterations specific to LDR and/or HDR samples, only alterations those could show 20× read coverage or more, were adopted because it was suggested that from 20× coverage onward, the minimum true mutation rate would be ≥97%, whereas the maximum false mutation started to reach a negligible level [9,11]. For amplicon sequencing, analyzed 50 genes and hotpots are described in the web site of CloneTech/Takara.
Results and Discussion
Candidates of Genome Alterations by WGS
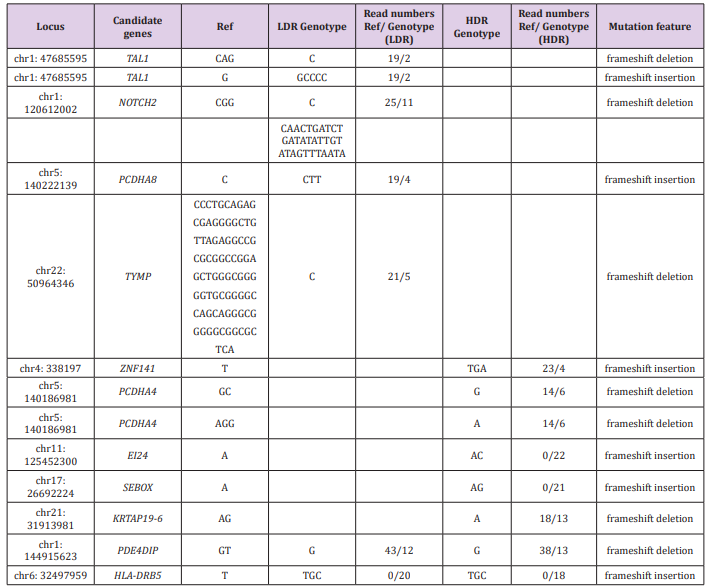
Table 1 shows genomic alterations in the LDR and/or HDR samples that were not present in the control sample. Six loci in four genes were found only in the LDR sample, six loci in five genes were found only in the HDR sample, and two loci in two genes were found in both samples. CAG>C and G>GCCCC alterations in the TAL1 gene, the CGG>C alteration in the NOTCH2 gene, C>CAACTGATCTGATATATTGTATAGTTTAATA and the C>CTT alteration in the PCDHA8 gene, the CCCTGCAGAGCGAGGGGCTGTTAGAGGCCGCGCGGCCGGAGCTGGGC GGGGGTGCGGGGCCAGCAGGGCGGGGGCGGCGCTCA>C alteration in the TYMP gene are the candidates of LDR-specific alterations. Because these alterations are frameshift or stop-gain mutations, production of proteins from these genes would be affected drastically. Among them, some genes are related to cancer development. Regulation of TAL1 gene expression was linked to T-cell acute lymphoblastic leukemia [12,13]. Targeting to NOTCH2 signaling promotes dedifferentiation of esophageal squamous cell carcinoma and breast cancer cells [14,15] and inhibits infiltration of glioblastoma cells [16]. The TYMP gene is linked to rectal cancer [17]. Regarding PCDHA8, ZNF141, PCDHA4, EI24, SEBOX, and HLA-DRB5 genes, their relationship with radiation and cancer development is unknown.
Table 1: Genomic alterations found in LDR and/or HDR samples compared to the reference sequences.
Note: Locus: Locations of the genome that showed alterations from the reference sequences (hg19), Candidate genes: Abbreviated gene names those showed alterations from the reference sequences (full names are described below), Ref: Bases of the hot spots of the reference sequences, LDR Genotype: Called genotypes in the LDR sample, Read numbers Ref/Genotype (LDR): Read numbers of the reference type/LDR Genotype, HDR Genotype: Called genotypes in the HDR sample, Read numbers Ref/Genotype (HDR): Read numbers of the reference type/HDR Genotype, Mutation feature: mutation feature type.
TAL1: T-cell acute lymphocytic leukemia protein 1, NOTCH2: Neurogenic locus notch homolog protein 2, PCDHA8: Protocadherin Alpha 8, TYMP : Thymidine Phosphorylase, ZNF141: Zinc Finger Protein 141, PCDHA4 : Protocadherin Alpha 4, EI24 : Autophagy Associated Transmembrane Protein, SEBOX: Skin-embryo-brain-oocyte homeobox, KRTAP19-6: Keratin Associated Protein 19-6, PDE4DIP: Phosphodiesterase 4D Interacting Protein, HLA-DRB5: Major Histocompatibility Complex, Class II, DR Beta 5.
Candidates of Genome Alterations by Amplicon Sequence
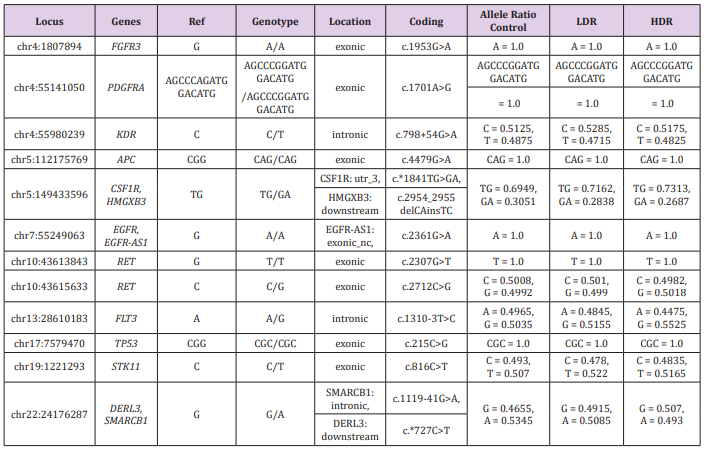
Because detection limits by WGS are usually in the range of 5-10% to as low as 1-2% [18], important alterations might be missed with the coverage of less than 20×. We decided to read specific cancer-related genes with deep coverage (more than 2,000× reads), because shallow reads in WGS might overlook LDR-specific alteration of genomic sequences. The “hotspot” regions analyzed by amplicon sequencing are well known to play critical roles in carcinogenesis (Table 2). Our results showed a same pattern of 12 hotspots of 50 genes and similar allele ratio between three samples, even at the hotspots differing from the reference gene. Thus, the ionizing irradiation used in this study did not cause any effects in the 50 genes. However, similar research using human embryonic stem cells (hESCs) that are highly ionizing irradiation-sensitive showed alterations of the KIT gene by 1 Gy, high-dose irradiation [18]. Because the cancer “hotspot” libraries generated in this study typically included 207 amplicons with an average size of 163 base pairs each, they would cover a very small portion (approximately 0.001%) of the human genome. Therefore, further research is necessary to investigate more cancer-related genes, including those we found in the WGS data.
Table 2: Comparison of the hot spots differed from the reference sequences.
Note: Locus: Locations of hot spots that showed alterations from the reference sequences (hg19); Genotype: Called genotypes, Ref: Bases of the hot spots of the reference sequences; Genes: Abbreviated gene names of the hot spots (full names are described below); Location: Locations of the hot spots among the genes described as exonic, intronic, or downstream; Coding: Positions and base alterations of the hot spots; Allele Ratio: Ratio of the called base or sequence for each genotype among 2,000 reads of the control, LDR and HDR genomic DNA.
FGFR3: Fibroblast growth factor receptor 3, PDGFRA : Platelet Derived Growth Factor Receptor Alpha, KDR : kinase insertion domain receptor, APC : adenomatous polyposis coli, CSF1R: Colony stimulating factor 1 receptor, HMGXB3: HMG domain-containing protein 3, EGFR : Epidermal Growth Factor Receptor, EGFR-AS1: EGFR antisense RNA 1, RET: Rearranged during transfection, FLT3: FMS (FibroMyalgia Syndrome)-like tyrosine kinase 3, TP53: Transformation-related protein 53, STK11 : Serine/threonine kinase 11, DERL3: Derlin 3, SMARCB1: SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1.
Acknowledgment
This study was supported by the basic research grant of Osaka Prefecture University, Osaka, Japan and Institute of Life Health Studies, Osaka, Japan.
References
- Kalanxhi E, Dahle J (2012) Transcriptional responses in irradiated and bystander fibroblasts after low dose α-particle radiation. Int J Radiat Biol 88(10): 713-719.
- Tanaka, Y, Furuta M (2018) Determination of accurate dose of γirradiation causing low-dose hyper-radio-sensitivity by using human fibroblast cells. Biomed J Sci & Tech Res 10(3): 1-4.
- Fisher KE, Zhang L, Wang J, Smith GH, Newman S, (2016) Clinical validation and implementation of a targeted next-generation sequencing assay to detect somatic variants in non-small cell lung, melanoma, and gastrointestinal malignancies. J Mol Diagn 18(2): 299-315.
- Valero V 3rd, Saunders TJ, He J, Weiss MJ, Cameron JL, et al. (2016) Reliable detection of somatic mutations in fine needle aspirates of pancreatic cancer with next-generation sequencing: implications for surgical management. Ann Surg 263(1): 153-161.
- Goldenberg D, Russo M, Houser K, Crist H, Derr JB, et al. (2017) Altered molecular profile in thyroid cancers from patients affected by the Three Mile Island nuclear accident. Laryngoscope 127(3): S1-S9.
- Ito M, Miyata Y, Hirano S, Kimura S, Irisuna F, et al. (2017) Therapeutic strategies and genetic profile comparisons in small cell carcinoma and large cell neuroendocrine carcinoma of the lung using next-generation sequencing. Oncotarget 8(65): 108936-108945.
- Xu MD, Liu SL, Feng YZ, Liu Q, Shen M, et al. (2017) Genomic characteristics of pancreatic squamous cell carcinoma, an investigation by using high throughput sequencing after in-solution hybrid capture. Oncotarget 8(9): 14620-14635.
- Xu H, Wu X, Sun D, Li S, Zhang S, et al. (2018) SEGF: A novel method for gene fusion detection from single-end next-generation sequencing data. Genes (Basel) 9(7): 331.
- Nath N, Esche J, Müller J, Jensen LR, Port M, et al. (2018) Exome sequencing discloses ionizing-radiation-induced DNA variants in the genome of human gingiva fibroblasts. Health Phys 115(1): 151-160.
- Karthikeyan S, Bawa PS, Srinivasan S (2016) hg19K: addressing a significant lacuna in hg19-based variant calling. Mol Genet Genomic Med 5(1): 15-20.
- Besaratinia A, Li H, Yoon JI, Zheng A, Gao H, Tommasi S (2012) A highthroughput next-generation sequencing-based method for detecting the mutational fingerprint of carcinogens. Nucleic Acids Res 40(15): e116.
- Haider Z, Larsson P, Landfors M, Köhn L, Schmiegelow K, et al. (2019) An integrated transcriptome analysis in T-cell acute lymphoblastic leukemia links DNA methylation subgroups to dysregulated TAL1 and ANTP homeobox gene expression. Cancer Med 8(1): 311-324.
- Benyoucef A, Palii CG, Wang C, Porter CJ, Chu A, Dai F, et al. (2016) UTX inhibition as selective epigenetic therapy against TAL1-driven T-cell acute lymphoblastic leukemia. Genes Dev 30(5): 5085-5021.
- Wang C, Zhang W, Zhang L, Chen X, Liu F, et al. (2016) miR-146a-5p mediates epithelial-mesenchymal transition of oesophageal squamous cell carcinoma via targeting Notch2. Br J Cancer 115(12): 1548-1554.
- Lee GH, Yoo KC, An Y, Lee HJ, Lee M, et al. (2018) FYN promotes mesenchymal phenotypes of basal type breast cancer cells through STAT5/NOTCH2 signaling node. Oncogene 37(14): 1857-1868.
- Hwang E, Yoo KC, Kang SG, Kim RK, Cui YH, et al. (2016) PKCδ activated by c-MET enhances infiltration of human glioblastoma cells through NOTCH2 signaling. Oncotarget 7(4): 4890-4902.
- Huang MY, Wu CH, Huang CM, Chung FY, Huang CW, et al. (2013) DPYD, TYMS, TYMP, TK1, and TK2 genetic expressions as response markers in locally advanced rectal cancer patients treated with fluoropyrimidinebased chemoradiotherapy. Biomed Res Int, pp. 931028.
- Nguyen V, Panyutin IV, Panyutin IG, Neumann RD (2016) A genomic study of DNA alteration events caused by ionizing radiation in human embryonic stem cells via next-generation sequencing. Stem Cells Int, pp. 1346521.

 Research Article
Research Article