Abstract
Purpose: Growing knowledge supports the importance of microRNAs in cell growth regulation, differentiation, apoptosis, and tumorigenesis. We addressed the associations between miRNA variants and risk of urothelial bladder carcinoma (UBCa).
Subjects and Methods: Sixty-six cases with UBCa (33 pTa, 13 pT1, 20 pT2) of low and high tumor grade (21 and 45 cases, respectively), and 156 healthy controls were genotyped for miR-196a2 rs11614913(C>T), miR-146a rs2910164(C>G), and miR-499 rs3746444(A>G) using TaqMan genotyping assays. Patients were stratified according to tumor stage, tumor grade, and risk of recurrence for UBCa. We used the SNPStats (https://www.snpstats.net) to choose best interactive model(s) of inheritance in selected miRNAs.
Results: The rs3746444 SNP showed a potential association with risk of UBCa (odds ratio [OR]= 1.9; P= 0.012). Significant associations were found for rs3746444 (P= 0.013) in codominant models, and for rs11614913 (P= 0.048) in an additive model. The rs11614913T allele had a protective effect, while homozygotes for the C allele were associated with a significantly increased risk of UBCa compared with controls (χ2= 3.7; P= 0.05). Homozygosity of rs3746444A/A was more frequent in cases than controls (43.9% versus 30.8%). Despite its significant impact in diverse ethnic populations, the rs29101164 SNP did not show clear effects on UBCa. Besides, the presence of rs3746444A/A was associated with tumors of high grade (OR= 2.7; P= 0.046).
Conclusion: This study was the first among the Saudi community to present a substantial function of miRNA biomarkers to predict UBCa, identify at-risk patients, and suggest a potential therapeutic target for UBCa.
Keywords: MicroRNAs; Urothelial Carcinoma of Bladder; Biomarkers; Genotyping
Abbreviations: BCG: Bacillus Calmette- Guérin; miRNA: microRNA; PCR: Polymerase Chain Reaction; SNP: Single Nucleotide Polymorphism; TaqMan Real-Time PCR; TNM: Tumor, Node, And Metastasis; TURBT: Transurethral Resection of Bladder Tumors; UBa; Urinary Bladder; UBCa: Urothelial Bladder Carcinoma
Introduction
Bladder cancer (BCa) is one of the most frequent malignancies worldwide. It has been characterized by a high recurrence rate, thus reflecting a substantial public health burden. In Saudi Arabia, almost 83% of all diagnosed BCa is urothelial bladder carcinoma (UBCa), and one-third of UBCa is described as invasive UBCa with a high risk for distant metastases [1]. Approximately 70% to 80% of patients are diagnosed with non-invasive UBCa (pTa-pT1), and the remaining patients have invasive UBCa that often leads to recurrence [2]. The recurrence rate within five years following the transurethral resection of bladder tumors (TURBT) can be estimated as 75% [3]. Thus, the disease may invade the muscle layer very rapidly. UBCa is not easy to cure because of its high recurrence and metastasis rates, with a five-year survival rate of about 57%. [4]. The great impact of microRNA (miRNA) has revolutionized current cell biology and medical science. miRNAs are a group of highly conserved small noncoding RNA molecules with 18-24 nucleotides [5]. They can post-transcriptionally regulate gene expression by directly targeting messenger RNAs (mRNAs). miRNAs can bind to the 3` untranslated regions of mRNAs, resulting in the degradation or translational repression of mRNAs. Previous reports revealed that miRNAs are widely involved in various processes of cell proliferation, differentiation, and apoptosis [6-8]. However, miRNAs can function as either oncogenes or tumor suppressors in different carcinomas, including BCa [6,9-17]. Reports on candidate genes and their impact on the risk of UBCa are still inadequate in Saudi population. Recently, we investigated associations between combinations of genetic variants of glutathione transferases, cytochrome P450, TP53 and MTHFR, and MTRR genes with the risk of UBCa among Saudi patients [18]. Here, we have extended our work to examine the effect of single nucleotide polymorphisms (SNPs) in miRNAs— the rs2910164 SNP in miR-146a, the rs11614913 SNP in miR-196a2, and the rs3746444 SNP in miR-499 genes—on the risk of UBCa in Saudi patients.
Subjects and Methods
Ethics Statement and Consent
The Institutional Biomedical Ethics Committee of Medicine College at Umm Al-Qura University approved the study protocols (reference #HAPO-02-K-012), licensed from the National Committee of Medical & Bioethics, KACST, Riyadh (http://bioethics. kacst.edu.sa/About.aspx?lang=en-US). All study participants signed an informed consent form.
Study Population
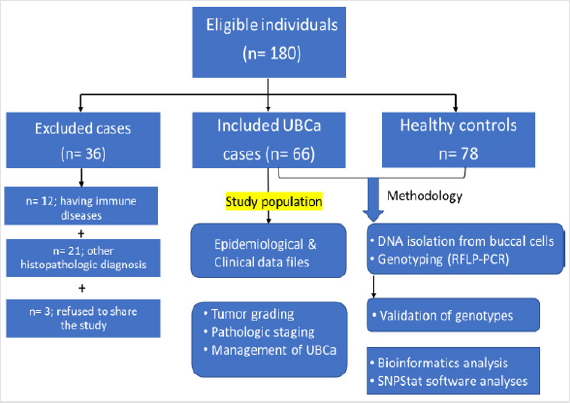
The study included 66 patients (ages 39-94 years) diagnosed with UBCa who were referred to the Urology Department at King Abdullah City Hospital (Mecca city) between June 2014 and January 2017. Epidemiologic and clinical characteristics regarding sex, age at examination, family history of cancer, cigarette smoking habits, alcohol consumption, pathologic tumor stage, tumor grade, and metastasis were recorded for each patient for statistical analyses (Figure 1). Patients were stratified according to: 1) pathological tumor staging using the tumor, node, and metastasis (TNM) classification system (pTa, pT1, pT2), 2) tumor grades according to the 2014 World Health Organization grading system,[19] and 3) recurrence risk according to the European Organization for Research and Treatment of Cancer risk criteria [20]. In the TNM system, ‘pTa’ is defined as a noninvasive papillary urothelial neoplasm of low malignancy, and the small section of tissue can be easily removed with TURBT. In contrast, ‘pT1’ describes a tumor that invades the subepithelial connective muscle but does not involve the bladder, and ‘pT2’ describes a tumor that has spread to the muscle of the bladder wall.
Figure 1: Flow chart of the eligible sample individuals, and applied methodology in the study population.
The management of the disease—through intravesical bacillus Calmette-Guérin (BCG), immunotherapy, a conservative TURBT‒ BCG therapy (injection), chemotherapy, chemoradiotherapy, or radical cystectomy—was also recorded. After TURBT was performed, the high-grade tumor cases were treated with BCG. Patients who previously had cancer, radiotherapy, and chemotherapy, or metastasized cancer from non-Saudi or unknown origins were excluded. Patients (n = 36) with immune disease or any histopathologic diagnosis other than UBCa were also excluded. Healthy individuals (n = 78) having no evidence of any clinical phenotypes of malignancies were selected as controls (ages 44-89 years) in routine follow-up at governmental hospitals in Makkah (Figure 1).
TaqMan Genotyping Analysis
Genomic DNA was extracted from peripheral blood (200 μL) using the QIAamp DNA blood kit as recommended by the manufacturer (Qiagen, Hilden, GmbH, Germany). In some cases, DNA samples were taken from buccal mucosa using the Oragene. DNA-OGR-575 kit (DNA Genotek Inc., Ottawa, ON, Canada) with some modifications [21]. We adopted TaqMan real-Time PCR analysis (Thermo, Applied Biosystems, USA) to genotype individuals for the selected SNPs of the miRNA 164a (rs2910164; C__15946974_10), miRNA 196a2 (rs11614913; C__31185852_10), and miRNA 499a (rs3746444; C___2142612_40) using a 7500 Fast-Dx Real-Time PCR System (Thermo, Applied Biosystems, Life Technologies Inc., USA). To ensure the accuracy of genotyping, all DNA samples, as well as negative controls, were included in the assays. We repeatedly genotyped 10% of the samples, and the results were 100% concordant. Some genotypes were validated by genotyping using a Genetic Analyzer 3500 (ABI, Life Technologies, Jeddah, Saudi Arabia).
Statistical Analysis
SNPs from all participants were tested for Hardy-Weinberg equilibrium (HWE) using the χ2 test, and a P value > 0.05 was considered consistent with HWE. We conducted the statistical analysis considering the interactive models of inheritance— codominant, dominant, recessive, over dominant, and additive— using the SNPStats software (https://www.snpstats.net). Logistic regressions for genotypic distributions and allelic frequencies for UBCa cases and controls were measured in terms of odds ratios (ORs) and 95% confidence intervals (CIs). The less Akaike information criterion (AIC) value that corresponded to the minimally expected entropy was adopted to assess the best model of inheritance. We used the MedCalc statistical software version 16.4.3 (MedCalc Software bvba, Ostend, Belgium; https://www.medcalc.org) to perform 𝑡-tests and chi-square tests for epidemiologic and clinical characteristics. A two-tailed P < 0.05 was considered statistically significant.
Result
Characteristics of the Study Population
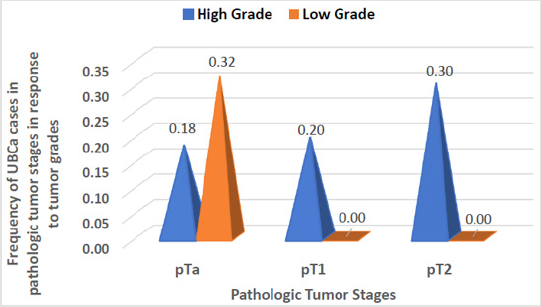
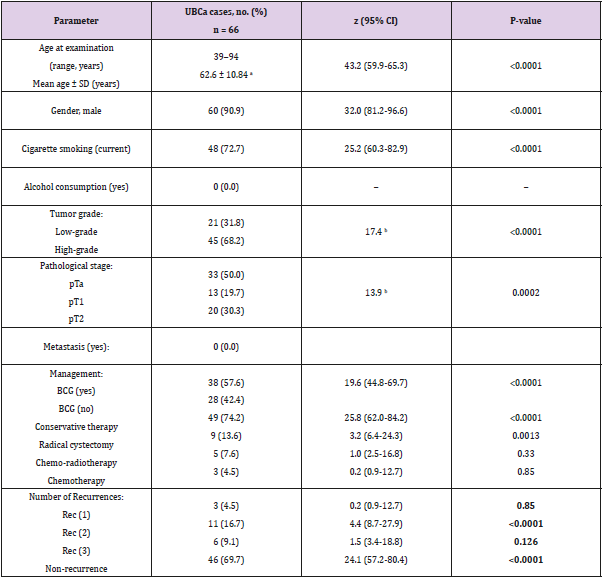
Sixty-six eligible Saudi individuals with UBCa (60 men, 6 women; 10:1 ratio) and 78 healthy controls (70 men, 8 women; 8.75:1 ratio) were enrolled. The mean age of patients was 62.6 ± 10.84 years, with no significant difference when compared with controls (t = 0.4, P = 0.7). There was no significant difference between cases and controls in terms of the percentage of current cigarette smokers (72.7% versus 71.8%, respectively; χ2 = 0.014, P = 0.9). A significant difference between the proportion of cases with a high tumor grade (45 cases, 68.2%) and the proportion with a low tumor grade (21 cases, 31.8%) was found (χ2 = 17.4, 95% CI 19.5-50.5; P < 0.0001). The percentages of UBCa cases with specific tumor stages were 50.0% for pTa, 19.7% for pT1, and 30.3% for pT2 (χ2 = 13.9; P = 0.0002) (Table 1). Among all cases, 18% (12 cases) with pTa, 20% (13 cases) with pT1, and 30% (20 cases) with pT2 tumors had high-grade tumors. No cases with pT1 or pT2 tumors had lowgrade tumors, and 21 (32%) of cases with milder pTa tumors had low-grade tumors (Figure 2). However, there was a highly significant difference between pathologic tumor stages with response to tumor grades among our cases with UBCa (χ2 = 30.8, P ≤ 0.001). The predominant management course for our cases with UBCa was BCG immunotherapy (76.9%; z = 23.8, P < 0.0001). The proportion of cases managed with TURBT-BCG injection (conservative therapy) was higher than the proportions managed with other strategies (74.2% versus 13.6% for radical cystectomy, 7.6% for chemo-radiotherapy, and 4.5% for chemotherapy) (Table 1).
Figure 2: Pathological stages and tumor grades in cases with urothelial bladder carcinoma (UBCa). The number of cases with tumors of “high grade” increased from pTa to pT2. There were no cases with tumors of “low grade” linked to pTa or pT2.
Table 1: Epidemiologic and clinical characteristics in UBCa cases.
Note: BCG: bacillus Calmette-Guérin; a weakened bacterium intravesically introduced via a catheter, Conservative therapy: TURBT
plus BCG, CI: confidence interval, SD: standard deviation, UBCa: urothelial bladder carcinoma, pTa, no invasive papillary carcinoma
and the small section of tissue can be easily removed with TURBT, pT1: tumor invades the subepithelial connective muscle but does
not involve the bladder, pT2: tumor has spread to the muscle of the bladder wall. Bold numbers indicate statistically significant
associations (P < 0.05). Rec (1, 2, 3) denotes the number of recurrences of UBCa.
aStudent’s t-test. Values are mean ± SD
bChi-square value.
HWE of SNPs in miRNA Regions
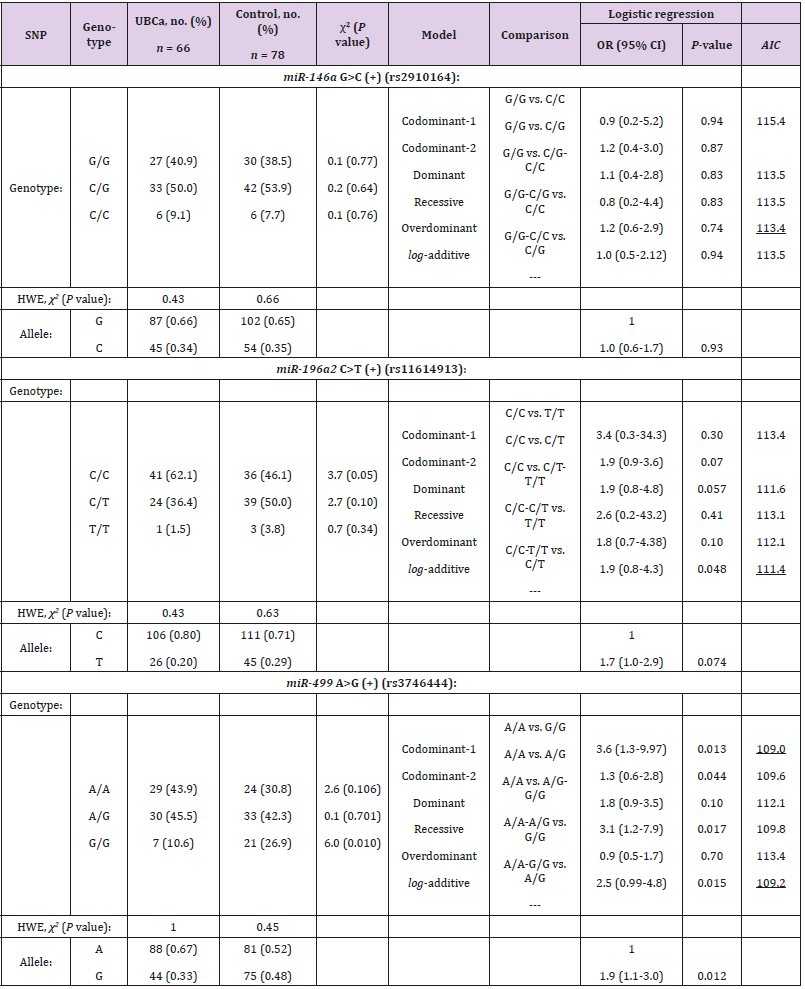
Three SNPs in the miRNAs’ regions—rs2910164 G>C, rs11614913 C>T, and rs3746444 A>G—were successfully genotyped in 66 patients with UBCa and 78 control subjects. All cases and controls were in HWE at the examined SNPs in the miRNAs (P > 0.05) (Table 2).
Table 2: Genotype distribution and allele frequencies of selected SNPs in microRNA regions and their associations with UBCa risk among cases and control.
Note: UBCa: urothelial bladder carcinoma, SNP: single nucleotide polymorphism, HWE: Hardy-Weinberg Equilibrium, OR: odds ratio, CI: confidence interval. AIC values refer to the model with the less AIC value that corresponds to the minimal expected entropy. Bold numbers indicate statistically significant associations (P < 0.05). Underlined numbers represent the best model of inheritance with the less AIC value.
Allele Frequencies and Interactive Genotypic Models
Regarding allele frequencies, the ORs of the allelic variants were 1.0 (95% CI, 0.6-1.7) for rs2910164, 1.7 (95% CI, 1.0-2.9) for rs11614913, and 1.9 (95% CI, 1.1-3.0) for rs3746444 (Table 2). However, the rs11614913C and rs3746444A alleles were more frequent in cases than in controls (0.80 versus 0.71; P = 0.074, and 0.67 versus 0.52; P = 0.012, respectively). In contrast, neither G allele of the rs2910164 SNP in cases or controls showed a clear effect on UBCa (0.66 versus 0.65, P = 0.93). In Table 2, the best interactive model of inheritance, described with the less AIC value, corresponds to the minimal expected entropy. Thus, evidence suggested that the best model was over dominant for rs2910164 (OR = 1.2; 95% CI 0.6-2.9; P = 0.74) and codominant-1 for rs11614913 and rs3746444 (OR = 1.9; 95% CI 0.8-4.3; P = 0.048, and OR = 3.6; 95% CI 1.3-9.97; P = 0.013, respectively). With slightly larger AIC values, the examined rs3746444 SNP showed that the codominant-2 (OR = 1.3; 95% CI 0.6-2.8; P = 0.044), recessive (OR = 3.1; 95% CI 3.1-7.9; P = 0.017), and log-additive (OR = 2.5; 95% CI 0.99-4.8; P = 0.015) models of inheritance were significantly associated with risk of UBCa. Consequently, examination of the rs3746444 SNP suggested an apparent role for homozygosity (codominant-1 model) in increasing risk for cases with UBCa compared to controls (43.9% versus 30.8%). As for rs11614913 C>T, the additive model reflected that each copy of T modified the protective effect in cases (additive model, 2T/T+C/T to C/C).
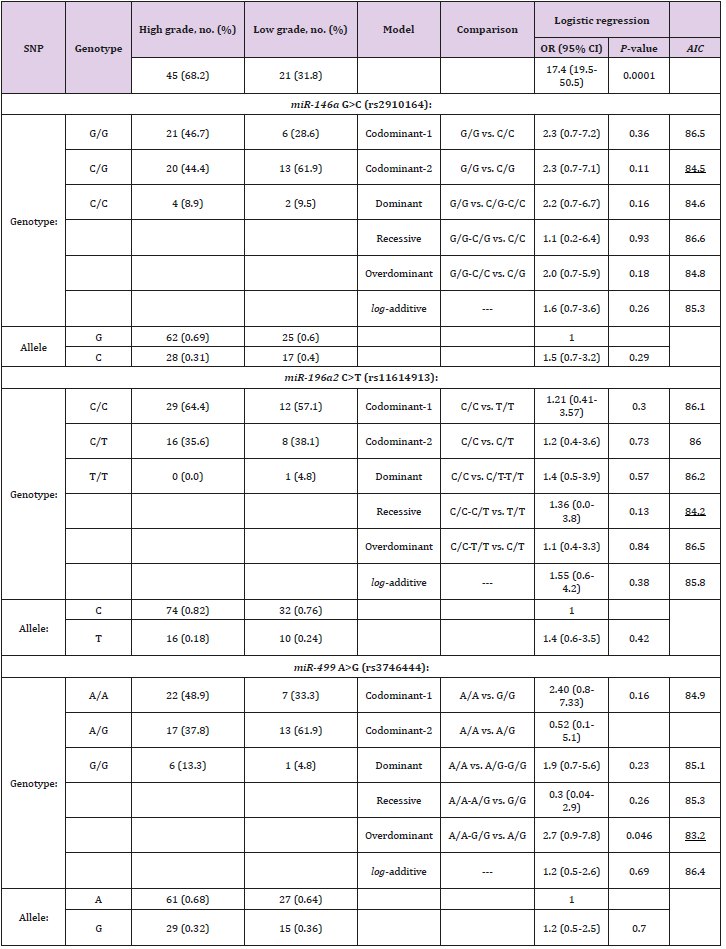
Genotyping associations of miRNAs and tumor grades
The interactive genotypic models of the selected SNPs in miRNAs with the response of tumor grades after adjusting for age and gender are shown in Table 3. There was a significant difference between the tumor grades of cases with UBCa in rs3746444 A>G in an overdominant model (OR = 2.7; 95% CI 0.9-7.8; P = 0.046), but not in rs2910164 (OR = 2.3; 95% CI 0.7-7.1; P = 0.11) or rs11614913 (OR = 1.4; 95% CI 0.0-3.8; P = 0.13). The proportions of the homozygotes in C/C in rs11614913 and A/A in rs3746444 were much higher in cases with high-grade tumors than those with low-grade tumors (62.1% versus 46.1%, and 43.9% versus 30.8%, respectively).
Table 3: Genotypic associations between selected SNPs in miRNAs and tumor grades among cases with UBCa.
Note: UBCa: urothelial bladder carcinoma, SNP: single nucleotide polymorphism, OR: odds ratio, CI: confidence interval. AIC values refer to the model with the less AIC value that corresponds to the minimal expected entropy. Bold numbers indicate statistically significant associations (P < 0.05). Underlined numbers represent the best model of inheritance with the less AIC value.
Discussion
This hospital-based case-control study presents the first evidence that common SNPs in miRNAs regions may be used as candidate genetic markers for UBCa vulnerability in a Saudi population. Our results show that the miR-499 rs3746444 SNP, but not the miR-164a rs2910164 or miR-196a2 rs11614913 SNP, is associated with risk for UBCa. Meta-analyses have been inconclusive regarding the association between the rs2910164, rs11614913, and rs3746444 SNPs in the miRNA genes and cancer risk [22-24]. Several reports have provided no evidence of an association between rs2910164 and overall cancer risk in diverse populations [22,23,25]. In our Saudi cases, the lack of association between the rs2910164 SNP and UBCa is consistent with previous results in Caucasian and Asian populations [26-28]. In addition, for the rs2910164C allele, reduced risks have been found for various types of cancer and in variable ethnic populations: cervical cancer in Chinese populations;[29] prostate in Iranian;[30] liver in Chinese, [31-34] Egyptian, [35] and Turkish [36]; colorectal cancer in Czech [37] and South Korean [38]; and gastric in Japanese [39] and Caucasian [40].
In contrast, the rs2910164C variant was found to be associated with increased risks of developing lung cancer, [41] and of developing head/neck cancer among Caucasian populations [42]. These discrepancies cannot be explained by variable ethnicity, but instead are likely due to organ-specific effects as well as different living environments, diets, climates, and lifestyles. Some previous results have suggested that rs11614913 and rs3746444 SNPs confer susceptibility to UBCa risk among different ethnic populations. However, consistent with our study, Mittal et al. [43] reported no association between the rs11614913 SNP and BCa. For rs11614913, some previous studies reported a significantly increased risk of certain cancers when T/T for C/C or combined C/C-C/T was present: breast cancer, [44] gliomas, [45] prostate cancer, [46] lung cancer, [47] colorectal cancer, [48] and liver cancer in patients infected with hepatitis B virus [49]. In agreement with our study, a meta-analysis found significant associations between allele frequency or different genotypic models within miR-499 rs3746444 SNP and risk of BCa in studies among Chinese populations [23].
In contrast, several reports could not find any associations between the rs3746444 SNP and BCa, [50] but did find associations with other types of cancer, including colorectal, liver, gallbladder, and breast cancer [38,51-53]. Based on the Human Gene Mutation Database, [54] human related-disease genes can be categorized into cancer disease genes and Mendelian disease genes. Several miRNAs have been found to regulate cancer migration, invasion, metastasis, and growth, and could be the targets for cancer therapy in about 30% of cancer genes [15,24,55-58]. Unfortunately, few articles have dealt with the regulation of BCa cells by these oncogene-targeted miRNAs, including miR-940/INPP4A, miR-940/GSK3b, miR-146a- 3p/PTTG1, and miR-145/PAK1 [59-61]. However, Xiang et al.60 found that overexpression of miR-146a-3p could inhibit BCa progression by targeting PTTG1 (OMIM 604147)—an oncogene that is expressed in many tumors and is correlated with tumor size, TNM stage, lymphatic invasion, and distant metastasis of BCa. Besides, small interfering RNA and miRNA can serve to increase the sensitivity of BCa cells to chemotherapeutic drugs [62-64]. For instance, miR-164a, miR-196a2, and miR-499 have been shown to hinder chemotherapeutic resistance in BCa [65,66].
Limitations
Meta-analysis has revealed that ethnicity could affect the association of miRNA polymorphisms with cancer risk Qiu et al. [67] reported that the significance of the sample size (P = 0.02), but not cancer type (P = 0.89) or source of controls (P = 0.97), contributed to the source of heterogeneity. Pinning down the miRNA polymorphisms for UBCa has been difficult because of poor replication of studies. Firstly, some studies have had different admixed populations, different sources of controls (hospital-based or population-based), or very small sample sizes, which would lower the strength of the overall results. Secondly, various studies included different SNPs of miRNAs that were not consistent with HWE in either controls or cases, which may have given rise to biased results of positive or negative associations. Thirdly, in our study, inclusion of more subjects in a realistic time frame from a specific hospital or clinical center would have been challenging, but replication of our results through larger, multicentric investigations will be of interest. Also, the literature includes other pathological types of cancer (e.g., squamous carcinoma or adenocarcinoma) in addition to UBCa, which might have influenced the outcomes of associations of the genetic markers with BCa. However, the present study focused only on UBCa, which consequently strengthens the reliability of our outcomes of associations [68,69].
Conclusion
Our results present the first evidence that miRNAs have great potential for clinical use as diagnostic biomarkers for UBCa susceptibility in a Saudi population. Our data show that the miR-499 rs3746444 A>G SNP contributes to an increased susceptibility to UBCa, as compared with the miR-164a rs2910164 G>C or the miR- 196a2 rs11614913 C>T SNPs. Furthermore, increased expression of miR-499 rs3746444 is significantly associated with a stage of UBCa with high-grade tumors. Our results regarding the rs3746444 SNP support roles for homozygosity and heterozygosity in affecting risk for UBCa, as one copy of the allelic variant rs3746444G could be necessary to produce a considerable damaging effect. In contrast, each copy of the T allele in rs11614913 can double the protective effect on individuals. Even though we found no association between the variant genotypes of miR-146a and UBCa risk in our Saudi population, miR-146a could exert a prominent role in carcinogenesis via marginal significance of heterozygosity in the debate of Asian and Caucasian ethnic studies. The inconclusive studies of cancer risk and the susceptibility conferred by the examined miRNAs may be due not only to admixture ethnicities and different living environments, diets, climates or lifestyles but also to organ-specific effects. Based on the results of previous clinical trials, some miRNAs under investigation can be used as biomarkers for risk prevention and therapeutic intervention. Ongoing analyses of UBCa patient DNA using whole-exome sequencing are being performed to discover more cancer genes that are targeted predominantly by several miRNAs. Thus, the miRNA targets on cancer genes analyzed will help enhance the knowledge UBa cancer genes and therapeutic improvements in treating cancer.
Acknowledgment
The authors would like to thank all subjects for their participation in this study. We wish to thank the Faculty of Biotechnology, Modern Sciences and Arts University (MSA), October City, Giza, for allowing the MSc student (ENE) to participate in the molecular genetic analyses.
Ethics Approval and Consent to Participate
Published work complies with the guidelines for human studies and the research was conducted ethically in accordance with the World Medical Association Declaration of Helsinki. Subjects have given their written informed consents and the study protocol on human research was approved by the Institutional Biomedical Ethics Committee of Umm Al-Qura University based on the National Committee of Biomedical Ethics at King Abdulaziz City for Sciences and Technology (KACST) (http://bioethics.kacst.edu.sa/About. aspx?lang=en-US).
Conflict of Interest
The authors have no economic or any conflicts of interest exists.
Consent for Publication
Written informed consent was obtained from all study participants to publish the results.
Data Availability
The data sets analyzed during the current study are available from the corresponding author.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Authors’ Contributions
NAE, and AMN designed the research; AMN, ASB, HAS, SNE, and HS made the clinical investigations and managements; NAE, AHM, MTT, and IAS, SNE, and ENE performed the practical work; NAE, IAS, AHM, and ENE work for in-silico predictions and statistical analyses; All authors shared in writing the draft, and reviewing and approving the final manuscript.
References
- Kaufman DS, Shipley WU, Feldman AS (2009) Bladder cancer. Lancet 374(9685): 239-249.
- Siegel RL, Miller KD, Jemal A (2015) Cancer statistics. CA Cancer J Clin 65(1): 5-29.
- Pruthi RS, Baldwin N, Bhalani V, Wallen EM (2008) Conservative management of low risk superficial bladder tumors. J Urol 179(1): 87-90.
- Larre S, Catto JW, Cookson MS, Messing EM, Shariat SF, et al. (2013) Screening for bladder cancer: rationale, limitations, whom to target, and perspectives. Eur Urol 63(6): 1049-1058.
- Grosshans H, Filipowicz W (2008) Molecular biology: the expanding world of small RNAs. Nature 451(7177): 414-416.
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2): 281-297.
- Bartel DP, Chen CZ (2004) Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 5(5): 396-400.
- Landi D, Gemignani F, Naccarati A, Pardini B, Vodicka P, et al. (2008) Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis 29(3): 579-584.
- Hu Z, Chen J, Tian T, Zhou X, Gu H, et al. (2008) Genetic variants of miRNA sequences and non-small cell lung cancer survival. J Clin Invest 118(7): 2600-2608.
- Pineau P, Volinia S, Mc Junkin K, Marchio A, Battiston C, et al. (2010) miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA 107(1): 264-269.
- Fan C, Chen C, Wu D (2013) The association between common genetic variant of microRNA-499 and cancer susceptibility: a meta-analysis. Mol Biol Rep 40(4): 3389-3394.
- Farazi TA, Hoell JI, Morozov P, Tuschl T (2013) MicroRNAs in human cancer. Adv Exp Med Biol 774: 1-20.
- Guo Y, Ying L, Tian Y, Yang P, Zhu Y, et al. (2013) miR-144 downregulation increases bladder cancer cell proliferation by targeting EZH2 and regulating Wnt signaling. FEBS J 280(18): 4531-4538.
- Song T, Zhang X, Zhang L, Dong J, Cai W, et al. (2013) miR-708 promotes the development of bladder carcinoma via direct repression of Caspase-2. J Cancer Res Clin Oncol 139(7): 1189-1198.
- Zhao X, Li J, Huang S, Wan X, Luo H, et al. (2015) MiRNA-29c regulates cell growth and invasion by targeting CDK6 in bladder cancer. Am J Transl Res 7(8): 1382-1389.
- Enokida H, Yoshino H, Matsushita R, Nakagawa M (2016) The role of microRNAs in bladder cancer. Investig Clin Urol 57(1): 60-76.
- Zhang C, Ma X, Du J, Yao Z, Shi T, et al. (2016) MicroRNA-30a as a prognostic factor in urothelial carcinoma of bladder inhibits cellular malignancy by antagonising Notch1. BJU Int 118(4): 578-589.
- Elhawary NA, Nassir A, Saada H, Dannoun A, Qoqandi O, et al. (2017) Combined genetic biomarkers confer susceptibility to risk of urothelial bladder carcinoma in a Saudi population. Dis Markers 2017: 1474560.
- Mac Lennan GT, Kirkali Z, Cheng L (2007) Histologic grading of noninvasive papillary urothelial neoplasms. Eur Urol 51(4): 889-897.
- Babjuk M, Oosterlinck W, Sylvester R, Kaasinen E, Bohle A, et al. (2011) European Association of U. EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder, the 2011 update. Eur Urol 59(6): 997-1008.
- Elhawary NA, Tayeb MT, Sindi IA, Qutub N, Rashad M, et al. (2019) Genetic biomarkers predict susceptibility to autism spectrum disorder through interactive models of inheritance in a Saudi community. Cogent Biol 5(1).
- Nikolic ZZ, Savic Pavicevic DL, Vucic NL, Romac SP, Brajuskovic GN, et al. (2015) Association between a genetic variant in the hsa-Mir-146a gene and cancer risk: an updated meta-analysis. Public Health Genomics 18(5): 283-298.
- Wang YH, Hu HN, Weng H, Chen H, Luo CL, et al. (2017) Association between polymorphisms in microRNAs and risk of urological cancer: a meta-analysis based on 17,019 subjects. Front Physiol 8: 325.
- Zhang L, Zhang M, Wang H, Wang Y, Zhou J, et al. (2018) Comprehensive review of genetic association studies and meta-analysis on polymorphisms in microRNAs and urological neoplasms risk. Sci Rep 8(1): 3776.
- Catucci I, Yang R, Verderio P, Pizzamiglio S, Heesen L, et al. (2010) Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Hum Mutat 31(1): 1052-1057.
- Yang H, Dinney CP, Ye Y, Zhu Y, Grossman HB, et al. (2008) Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res 68(7): 2530-2537.
- Wang M, Chu H, Li P, Yuan L, Fu G, et al. (2012) Genetic variants in miRNAs predict bladder cancer risk and recurrence. Cancer Res 72(23): 6173-6182.
- Ma XP, Zhang T, Peng B, Yu L, Jiang de K, et al. (2013) Association between microRNA polymorphisms and cancer risk based on the findings of 66 case-control studies. PLoS One 8(11): 79584.
- Yue C, Wang M, Ding B, Wang W, Fu S, et al. (2011) Polymorphism of the pre-miR-146a is associated with risk of cervical cancer in a Chinese population. Gynecol Oncol 122(1): 33-37.
- Hashemi M, Moradi N, Ziaee SA, Narouie B, Soltani MH, et al. (2016) Association between single nucleotide polymorphism in miR-499, miR-196a2, miR-146a and miR-149 and prostate cancer risk in a sample of Iranian population. J Adv Res 7(3): 491-498.
- Xu T, Zhu Y, Wei QK, Yuan Y, Zhou F, et al. (2008) A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis 29(11): 2126-2131.
- Zhou J, Lv R, Song X, Li D, Hu X, et al. (2012) Association between two genetic variants in miRNA and primary liver cancer risk in the Chinese population. DNA Cell Biol 31(4): 524-530.
- Chu YH, Hsieh MJ, Chiou HL, Liou YS, Yang CC, et al. (2014) MicroRNA gene polymorphisms and environmental factors increase patient susceptibility to hepatocellular carcinoma. PLoS One 9(2): 89930.
- Cong N, Chen H, Bu WZ, Li JP, Liu N, et al. (2014) miR-146a G>C polymorphisms and risk of hepatocellular carcinoma in a Chinese population. Tumour Biol 35(6): 5669-5673.
- Toraih EA, Fawz MS, Elgazzaz MG, Hussein MH, Shehata RH, et al. (2016) Combined genotype analyses of precursor miRNA196a2 and 499a variants with hepatic and renal cancer susceptibility a preliminary study. Asian Pac J Cancer Prev 17(7): 3369-3375.
- Akkiz H, Bayram S, Bekar A, Akgollu E, Uskudar O, et al. (2011) No association of pre-microRNA-146a rs2910164 polymorphism and risk of hepatocellular carcinoma development in Turkish population: A case-control study. Gene 486(1): 104-109.
- Hezova R, Kovarikova A, Bienertova Vasku J, Sachlova M, Redova M, et al. (2012) Evaluation of SNPs in miR-196-a2, miR-27a and miR-146a as risk factors of colorectal cancer. World J Gastroenterol 18(22): 2827-2831.
- Min KT, Kim JW, Jeon YJ, Jang MJ, Chong SY, et al. (2012) Association of the miR-146aC>G, 149C>T, 196a2C>T, and 499A>G polymorphisms with colorectal cancer in the Korean population. Mol Carcinog 51(1): 65-73.
- Parlayan C, Ikeda S, Sato N, Sawabe M, Muramatsu M, et al. (2014) Association analysis of single nucleotide polymorphisms in miR-146a and miR-196a2 on the prevalence of cancer in elderly Japanese: a case-control study. Asian Pac J Cancer Prev 15(5): 2101-2107.
- Kupcinskas J, Bruzaite I, Juzenas S, Gyvyte U, Jonaitis L, et al. (2014) Lack of association between miR-27a, miR-146a, miR-196a-2, miR-492 and miR-608 gene polymorphisms and colorectal cancer. Sci Rep 4: 5993.
- Lung RW, Wang X, Tong JH, Chau SL, Lau KM, et al. (2013) A single nucleotide polymorphism in microRNA-146a is associated with the risk for nasopharyngeal carcinoma. Mol Carcinog 52(1): 28-38.
- Niu YM, Du XY, Cai HX, Zhang C, Yuan RX, et al. (2015) Increased risks between Interleukin-10 gene polymorphisms and haplotype and head and neck cancer: a meta-analysis. Sci Rep 5: 17149.
- Mittal RD, Gangwar R, George GP, Mittal T, Kapoor R, et al. (2011) Investigative role of pre-microRNAs in bladder cancer patients: a case-control study in North India. DNA Cell Biol 30(6): 401-406.
- Hu Z, Liang J, Wang Z, Tian T, Zhou X, et al. (2009) Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat 30(1): 79-84.
- Dou T, Wu Q, Chen X, Ribas J, Ni X, et al. (2010) A polymorphism of microRNA196a genome region was associated with decreased risk of glioma in Chinese population. J Cancer Res Clin Oncol 136(12): 1853-1859.
- George GP, Gangwar R, Mandal RK, Sankhwar SN, Mittal RD, et al. (2011) Genetic variation in microRNA genes and prostate cancer risk in North Indian population. Mol Biol Rep 38(3): 1609-1615.
- Hong YS, Kang HJ, Kwak JY, Park BL, You CH, et al. (2011) Association between microRNA196a2 rs11614913 genotypes and the risk of non-small cell lung cancer in Korean population. J Prev Med Public Health 44(3): 125-130.
- Zhan JF, Chen LH, Chen ZX, Yuan YW, Xie GZ, et al. (2011) A functional variant in microRNA-196a2 is associated with susceptibility of colorectal cancer in a Chinese population. Arch Med Res 42(2): 144-148.
- Li XD, Li ZG, Song XX, Liu CF (2010) A variant in microRNA-196a2 is associated with susceptibility to hepatocellular carcinoma in Chinese patients with cirrhosis. Pathology 42(7): 669-673.
- Deng S, Wang W, Li X, Zhang P (2015) Common genetic polymorphisms in pre-microRNAs and risk of bladder cancer. World J Surg Oncol 13: 297.
- Cheng T, Wang L, Li Y, Huang C, Zeng L, et al. (2013) Differential microRNA expression in renal cell carcinoma. Oncol Lett 6(3): 769-776.
- Lv M, Dong W, Li L, Zhang L, Su X, et al. (2013) Association between genetic variants in pre-miRNA and colorectal cancer risk in a Chinese population. J Cancer Res Clin Oncol 139(8): 1405-1410.
- Vinci S, Gelmini S, Mancini I, Malentacchi F, Pazzagli M, et al. (2013) Genetic and epigenetic factors in regulation of microRNA in colorectal cancers. Methods 59(1): 138-146.
- Stenson PD, Ball EV, Mort M, Phillips AD, Shaw K, et al. (2012) The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr Protoc Bioinformatics 1: 13.
- Das J, Podder S, Ghosh TC (2014) Insights into the miRNA regulations in human disease genes. BMC Genomics 15: 1010.
- Xiaoli Z, Yawei W, Lianna L, Haifeng L, Hui Z, et al. (2015) Screening of target genes and regulatory function of miRNAs as prognostic indicators for prostate cancer. Med Sci Monit 21: 3748-3759.
- Wu K, Mu XY, Jiang JT, Tan MY, Wang RJ, et al. (2018) miRNA26a5p and miR26b5p inhibit the proliferation of bladder cancer cells by regulating PDCD10. Oncol Rep 40(6): 3523-3532.
- Zhou W, He L, Dai Y, Zhang Y, Wang J, et al. (2018) MicroRNA-124 inhibits cell proliferation, invasion and migration by targeting CAV1 in bladder cancer. Exp Ther Med 16(4): 2811-2820.
- Kou B, Gao Y, Du C, Shi Q, Xu S, et al. (2014) miR-145 inhibits invasion of bladder cancer cells by targeting PAK1. Urol Oncol 32(6): 846-854.
- Xiang W, Wu X, Huang C, Wang M, Zhao X, et al. (2017) PTTG1 regulated by miR-146a-3p promotes bladder cancer migration, invasion, metastasis and growth. Oncotarget 8(1): 664-678.
- Wang R, Wu Y, Huang W, Chen W (2018) MicroRNA-940 Targets INPP4A or GSK3beta and Activates the Wnt/beta-catenin pathway to regulate the malignant behavior of bladder cancer cells. Oncol Res 26(1): 145-155.
- Gandhi NS, Tekade RK, Chougule MB (2014) Nanocarrier mediated delivery of siRNA/miRNA in combination with chemotherapeutic agents for cancer therapy: current progress and advances. J Control Release 194: 238-256.
- Takai T, Yoshikawa Y, Inamoto T, Minami K, Taniguchi K, et al. (2017) A novel combination RNAi toward Warburg effect by replacement with miR-145 and silencing of PTBP1 induces apoptotic cell death in bladder cancer cells. Int J Mol Sci 18(1).
- Xiao J, Niu S, Zhu J, Lv L, Deng H, et al. (2018) miR223p enhances multi-chemoresistance by targeting NET1 in bladder cancer cells. Oncol Rep 39(6): 2731-2740.
- Bu Q, Fang Y, Cao Y, Chen Q, Liu Y, et al. (2014) Enforced expression of miR-101 enhances cisplatin sensitivity in human bladder cancer cells by modulating the cyclooxygenase-2 pathway. Mol Med Rep 10(4): 2203-2209.
- Pan J, Li X, Wu W, Xue M, Hou H, et al. (2016) Long non-coding RNA UCA1 promotes cisplatin/gemcitabine resistance through CREB modulating miR-196a-5p in bladder cancer cells. Cancer Lett 382(1): 64-76.
- Qiu MT, Hu JW, Ding XX, Yang X, Zhang Z, et al. (2012) Hsa-miR-499 rs3746444 polymorphism contributes to cancer risk: a meta-analysis of 12 studies. PLoS One 7(12): 50887.
- Shi HB, Yu JX, Yu JX, Feng Z, Zhang C, et al. (2017) Diagnostic significance of microRNAs as novel biomarkers for bladder cancer: a meta-analysis of ten articles. World J Surg Oncol 15(1): 147.
- Pollard J, Burns PA, Hughes TA, Ho Yen C, Jones JL, et al. (2018) Differential expression of microRNAs in breast cancers from four different ethnicities. Pathobiology 85(4): 220-226.

 Research Article
Research Article