Abstract
Background: Embelin is a major active compound of Embelia ribes, exerting a wide spectrum of pharmacological activities. However, the studies about molecular mechanisms induced by embelin have not been widely reported yet, most of which are not systematical and conclusive.
Methods: First, we evaluated the druggability of embelin using the Traditional Chinese Medicine Systems Pharmacology Database (TCMSP) server. Next, the potential target genes were predicted by chemical-gene interaction analysis. Furtherly, gene ontology and pathway analyses were investigated with the target genes. Finally, we constructed a drug-target-pathway network to provide a systematic overview of the potential target genes and the mechanisms of action about embelin.
Results: The pharmacokinetic properties of embelin meet all these requirements of Lipinski’s rule of five, meaning embelin could be considered as a preferable candidate for drug development. 31 target genes of embelin were identified and used to performed GO and KEGG analyses. GO, KEGG, and network analyses revealed that these targets were associated with cancers and hepatitis B. Notably, there were 9 pathways directly related to cancer among the top 10 predicted KEGG pathways, suggesting that embelin might exert dramatic effect on multiple cancers with diverse underlying mechanisms.
Conclusion: Embelin could be a promising compound for the development of effective multi-targeted drug, especially for treatment of cancers. Large-scale and comprehensive research is needed to clarify our results.
Highlights
1) The pharmacokinetic properties of embelin meets all the requirements of Lipinski’s rule of five.
2) Systematic analysis of mechanisms about embelin via network pharmacology.
3) Embelin, a promising compound for the development of effective multi-targeted drug, especially for treatment of cancers.
Keywords: Embelin; Network pharmacology; Druggability; Mechanism; Cancer; Disease
Abbreviations: TCM: Traditional Chinese Medicine; ADME: Absorption, Distribution, Metabolism, and Excretion; MW: Molecular Weight; AlogP: Lipid-Water Partition Coefficient Log P; Hdom: Hydrogen- Bond Donors; Hacc: Hydrogen-Bond Acceptors; OB: Oral Bioavailability; FASA- : Fractional Negative Accessible Surface Area; TPSA: Topological Polar Surface Area; RBN: Number of Rotatable Bonds; HL: Half Life; DL: Drug Likeness; Caco-2: Caco-2 Permeability; BBB: Blood-Brain Barrier; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; CTD: Comparative Toxicogenomics Database
Introduction
Natural products provide a resource of numerous active compounds for drug discovery. Identification and investigation of active constituent from natural plants are important for assessing their potential therapeutic effects. Embelia ribes has been widely used as an herb in traditional medicine in India and China for a long time [1]. Embelin (2,5-dihydroxy-3-undecyl-1,4-benzoquinone) is a major active compound of Embelia ribes, which belongs to the Myrsinaceae family. It could be also found in other plants, such as Radix Clematidis, Lysimachia punctata (Primulaceae) [2], and Erythrorhiza (Oxalidaceae) [3]. In recent decades, increasing evidence shows that embelin could exert multiple pharmacological activities, for example anticancer activity [4,5], anti-inflammatory activity [6], anthelmintic activity [7], antidiabetic property [8], neuroprotective effect [9,10], hepatoprotective effect [11] and so on. Embelin could be considered as a valuable compound with complex biological processes and pathways with diverse underlying mechanisms due to its effect in kinds of diseases [4].
However, the studies about molecular mechanisms induced by embelin and the resulting changes in cellular phenotypes have not been widely reported yet, most of which are not systematical and conclusive. Meanwhile, network pharmacology, which was firstly reported by Hopkins in 2007 [12], could provide a systematic overview of promising signaling pathways related to a certain compound [13]. Furthermore, target identification as well as the molecular mechanisms might be paid attention to in drug discovery and design processes. Therefore, we tried to reveal the effects of embelin by bioinformatics methods systematically. Firstly, we evaluated the druggability of embelin using TCMSP server [14]. Secondly, a chemical-gene interaction analysis was used to identify the potential target genes [15]. Furtherly, we performed gene ontology and pathway analyses with those target genes. Finally, we constructed a network including embelin, targets and pathways in order to provide an overview of the potential target genes and the mechanisms of function about embelin.
Materials and Methods
Assessment of Pharmacokinetics Properties
TCMSP database (http://lsp.nwu.edu.cn/tcmsp.php) is a database of systems pharmacology for traditional Chinese medicine (TCMs) and related compounds [14]. It was structured to incorporate several important absorption, distribution, metabolism, and excretion (ADME)-related features such as human oral bioavail-ability (OB), half-life (HL), drug-likeness (DL), fractional negative accessible surface area (FASA-), Caco-2 permeability (Caco-2), blood-brain barrier (BBB), number of rotatablebonds (RBN), opological polar surface area (TPSA) and Lipinski’s rule including molecular weight (MW), lipid-water partition coefficient log P (AlogP), hydrogen-bond donors (Hdon) and hydrogen-bond acceptors (Hacc)[16,17].
Target genes Identification
The Comparative Toxicogenomics Database (CTD, http:// ctdbase.org/), a publicly available database, could provide numerous information about chemical-gene-disease interactions relationships according to scientific literature [15]. Given a certain chemical, CTD would show the corresponding target genes immediately.
Analysis by GeneMania
GeneMANIA (http://www.genemania.org) could provide many bioinformatics services including predicting gene function, analyzing gene lists and prioritizing genes for functional assays [18]. With a gene list, GeneMANIA can prodict the proteins sharing properties or function similarly with the original content.
GO and KEGG Pathway Analyses
The web-based program Database for Annotation, Visualization and Integrated Discovery (DAVID, http:// david.abcc.ncifcrf.gov/) was used to perform functional enrichment analysis including Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and Gene Ontology (GO) annotation for the target genes [19-21]. A count number larger than 2 and P-value < 0.05 were chosen as cutoff criteria. Visualization of the results of GO and KEGG analyses was done using ggplot2 package of R software [22].
Network Construction
To understand the complex relationships among embelin, target genes and pathways, we chose Cytoscape (v 3.7.1) to construct and analyze the three-layer network.
Results
Pharmacokinetics Properties of Embelin
We use TCMSP to ensure AMDE-related properties of embelin including MW, AlogP, Hdom, Hacc, OB, Caco-2, BBB, DL, FSAF, TPSA, RBN and HL. The information was all shown in Table 1. Notably, the OB of embelin was 37.72%.
Targets Identification of Embelin
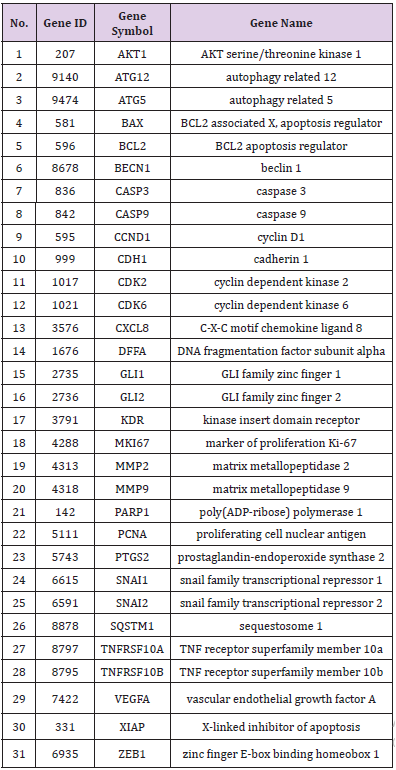
Totally, CTD identified 33 candidate target genes. Then, the 33 genes were filtered using the threshold chemical-gene interaction ≥1. Finally, 31 unique target genes for embelin remained (Table 2). Then further investigation was based on these 31 target genes.
GeneMania Analysis
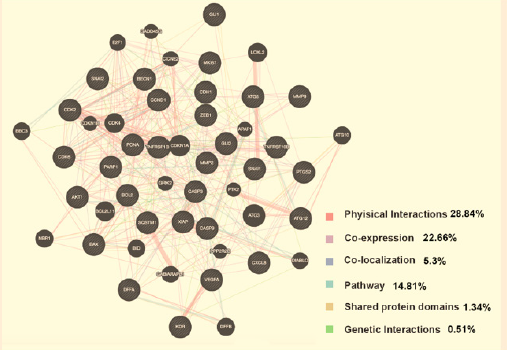
The 31 targets and their interacting proteins showed 28.84% physical interactions, 5.3% exerted co-localization and 22.66% similar co-expression characteristics. All the data above were provided as a network in Figures 1 & 2.
Figure 2: Protein network of embelin. Black nodes represent target proteins, and connecting colors indicate different correlations.
GO and Pathway Analyses
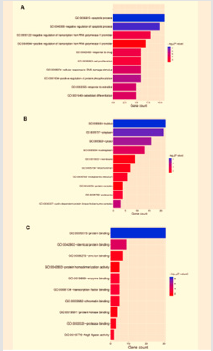
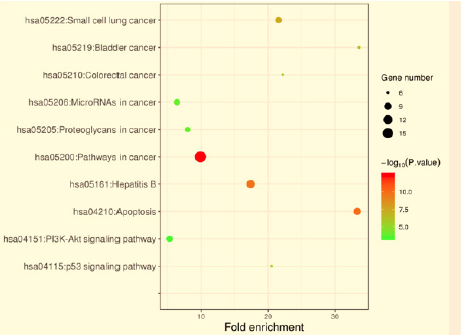
To study the 31 identified target genes furtherly, we performed GO and KEGG enrichment analyses using DAVID. As shown in Figure 3, the top seven functions by GO analysis were demonstrated as apoptotic process (11/31), negative regulation of apoptotic process (10/31), negative regulation of transcription from RNA polymerase II promoter (8/31), positive regulation of transcription from RNA polymerase II promoter (7/31), protein binding (31/31), identical protein binding (9/31), zinc ion binding (9/31). These functional terms were highly related to cell growth processes. As for KEGG analysis, the targets participated in top 10 pathways with gene counts adjusted P-value including “pathways in cancer”, “hepatitis B”, “apoptosis”, “small cell lung cancer”, “microRNAs in cancer”, “PI3K-Akt signaling pathway”, “proteoglycans in cancer”, “bladder cancer”, “colorectal cancer”, and “p53 signaling pathway”.
Figure 3: GO analysis (top 10) of putative target genes.
(A) Biological process categories.
(B) Cellular component categories.
(C) Molecular function categories.
Network Analysis
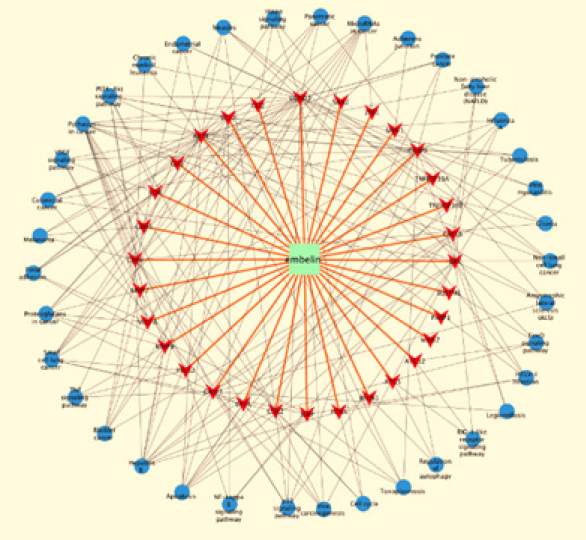
Based on the complex relationships among embelin, target genes and pathways, we constructed an entire compound-targetpathway network by Cytoscape. As shown in Figure 4, this network has 60 nodes and 226 edges. The green oblong, red inverted triangles, and blue circles correspond to embelin, target genes and pathways, respectively.
Discussion
Development of new therapeutic agents from natural sources has great promise for diverse diseases, especially human cancers [23]. Network pharmacology approach to systematic and multitarget drug discovery could lead to a new generation of candidates with improved physicochemical and pharmacokinetics properties [24,25]. Today, Lipinski’s rule of five is generally known as a criterion for drug optimization. The important properties involved in the rule should be considered for oral drugs discovery [26]. The rule of five means MW < 500 Da, ALogP < 5, as well as numbers of Hdom and Hacc less than 5 and 10, respectively. Moreover, among all pharmacokinetics properties, OB is the most important feature for orally administered drugs [14,16]. The pharmacokinetic properties of embelin met all the requirements of Lipinski’s rule of five (Table 1), meaning that embelin could be considered as a preferable candidate for drug development. As is an important step in drug discovery, target identification investigates the mechanisms of effects about compounds by identifying their interacting proteins [27-30] (Graphic Abstract).
Kinds of approaches have been developed and applied widely. As listed in Table 2, 31 potential targets of embelin were identified using CTD. GeneMANIA provided information on physical interactions, co-localization, co-expression as well as shared protein domains. The website also implied that the targets and their interacting proteins may possess specific or similar functions. Several of the potential targets of embelin have been reported in other studies. For example, embelin induced autophagic and apoptotic cell death in human oral squamous cell carcinoma cells via regulating ATG12/ATG5 complex [31]. Thus, the therapeutic effects of enbelin might be mediated by multiple targets, often interacting synergistically. Next, GO and KEGG analyses were conducted. As shown in Figures 3 & 5, embelin had multiple targets and therefore exerted multiple pharmacological effects. Interestingly, there were 9 pathways directly related to cancer among the top 10 significant KEGG pathways (9/10) predicted from DAVID.
The 9 pathways included “pathways in cancer”, “apoptosis”, “small cell lung cancer”, “microRNAs in cancer”, “PI3K-Akt signaling pathway”, “proteoglycans in cancer”, “bladder cancer”, “colorectal cancer”, and “p53 signaling pathway”, suggesting that embelin might exert dramatic effect on multiple cancers with diverse underlying mechanisms. Previous studies have reported that embelin was effective against cancers. For instance, in lung cancer cells, activation of p53 was found to play a pivotal role in the apoptotic activity of embelin [32]. Moreover, treatment of lung cancer cells with embelin caused activation of p-p38 and p-JNK, which led to the activation of caspase-3 and cell death [33]. Additionally, embelin inhibited growth of bladder cancer cells by inducing apoptosis via PI3K/Akt pathway [34]. For the rest pathway related to hepatitis B, embelin has been explored as one of novel hepatitis B virus inhibitors [35]. These findings above closely coincide with our results from GO and KEGG analyses. The drug-target-pathway network (Figure 4) also revealed that embelin had multiple targets and it could possess multiple pharmacological effects, consist with several well-known studies [1]. Embelin are more effective for the treatment of complex diseases, for instance, hepatitis B and tumors. Hence, embelin could be a promising compound that might be an active candidate for future drug discovery in treatment of numbers of diseases, especially human cancers.
In conclusion, we would like to underline that embelin could be a promising compound for the development of a safe and effective multi-targeted drug. It is also hoped that this study would encourage further research on the pharmacological effects of embelin, both in vitro and in vivo, in order to provide a more in-depth insight into the mechanisms associated with the therapeutic effects of embelin.
Acknowledgement
This work was financially supported by Natural Science Foundation of China (81502294) and the Project of Jiangsu Province Traditional Chinese Medicine Bureau (YB201952).
Conflict of Interest
The authors declare no conflict of interest.
References
- Lu H, Wang J, Wang Y, Qiao L, Zhou Y (2016) Embelin and Its Role in Chronic Diseases. Advances in experimental medicine and biology 928: 397-418.
- Podolak I, Galanty A, Janeczko Z (2005) Cytotoxic activity of embelin from Lysimachia punctate. Fitoterapia 76(3-4): 333-335.
- GE Feresin, A Tapia, M Sortino, S Zacchino, AR de Arias, et al. (2003) Bioactive alkyl phenols and embelin from Oxalis erythrorhiza. Journal of ethnopharmacology 88: 241-247.
- Ko JH, Lee SG, Yang WM, Um JY, Sethi G, et al. (2018) The Application of Embelin for Cancer Prevention and Therapy. Molecules (Basel, Switzerland) 23(3).
- Prabhu KS, Achkar IW, Kuttikrishnan S, Akhtar S, Khan AQ, et al. (2018) Embelin: a benzoquinone possesses therapeutic potential for the treatment of human cancer. Future medicinal chemistry 10(8): 961-976.
- Kalyan Kumar G, Dhamotharan R, Kulkarni NM, Mahat MY, Gunasekaran J, et al. (2011) Embelin reduces cutaneous TNF-alpha level and ameliorates skin edema in acute and chronic model of skin inflammation in mice. European journal of pharmacology 662(1-2): 63-69.
- D'Avigdor E, Wohlmuth H, Asfaw Z, Awas T (2014) The current status of knowledge of herbal medicine and medicinal plants in Fiche, Ethiopia. Journal of ethnobiology and ethnomedicine 10: 38.
- Bhandari U, Jain N, Pillai KK (2007) Further studies on antioxidant potential and protection of pancreatic beta-cells by Embelia ribes in experimental diabetes. Experimental diabetes research pp. 15803.
- Bhuvanendran S, Hanapi NA, Ahemad N, Othman I, Yusof SR, et al. (2019) A Potent Molecule for Alzheimer's Disease: A Proof of Concept From Blood-Brain Barrier Permeability, Acetylcholinesterase Inhibition and Molecular Docking Studies. Frontiers in neuroscience 13: 495.
- UP Kundap, S Bhuvanendran, Y Kumari, I Othman, MF Shaikh (2017) Plant Derived Phytocompound, Embelin in CNS Disorders: A Systematic Review. Frontiers in pharmacology 8: 76.
- Sreepriya M, Bali G (2007) Effects of administration of Embelin and Curcumin on lipid peroxidation, hepatic glutathione antioxidant defense and hematopoietic system during N-nitrosodiethylamine/Phenobarbital-induced hepatocarcinogenesis in Wistar rats. Molecular and cellular biochemistry 284(1-2): 49-55.
- AL Hopkins (2007) Network pharmacology Nature biotechnology 25: 1110-1111.
- Chen SJ, Cui MC (2017) Systematic Understanding of the Mechanism of Salvianolic Acid A via Computational Target Fishing. Molecules (Basel, Switzerland) 22(4).
- Ru J, Li P, Wang J, Zhou W, Li B, et al. (2014) TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. Journal of cheminformatics 6: 13.
- AP Davis, CJ Grondin, RJ Johnson, D Sciaky, R McMorran, et al. (2019) The Comparative Toxicogenomics Database: update 2019. Nucleic acids research 47: D948-D954.
- Pei T, Zheng C, Huang C, Chen X, Guo Z, et al. (2016) Systematic understanding the mechanisms of vitiligo pathogenesis and its treatment by Qubaibabuqi formula. Journal of ethnopharmacology 190: 272-287.
- Zhang W, Tao Q, Guo Z, Fu Y, Chen X, et al. (2016) Systems Pharmacology Dissection of the Integrated Treatment for Cardiovascular and Gastrointestinal Disorders by Traditional Chinese Medicine. Scientific reports 6: 32400.
- Warde Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, et al. (2010) The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic acids research 38: W214-220.
- DW Huang, BT Sherman, Q Tan, J Kir, D Liu, et al. (2007) DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic acids research 35: W169-175.
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, et al. (2008) KEGG for linking genomes to life and the environment. Nucleic acids research 36: D480-484.
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics 25(1): 25-29.
- Ito K, Murphy D (2013) Application of ggplot2 to Pharmacometric Graphics. CPT: Pharmacometrics & systems pharmacology 2: e79.
- H van de Waterbeemd, E Gifford (2003) ADMET in silico modelling: towards prediction paradise? Nature reviews. Drug discovery 2: 192-204.
- YH Bi, LH Zhang, SJ Chen, QZ Ling (2018) Antitumor Mechanisms of Curcumae Rhizoma Based on Network Pharmacology. Evidence-based complementary and alternative medicine pp. 4509892.
- Lin YC, Chang CW, Wu CR (2015) Anti-nociceptive, anti-inflammatory and toxicological evaluation of Fang-Ji-Huang-Qi-Tang in rodents. BMC complementary and alternative medicine 15: 10.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced drug delivery reviews 46(1-3): 3-26.
- M Schenone, V Dancik, BK Wagner, PA Clemons (2013) Target identification and mechanism of action in chemical biology and drug discovery. Nature chemical biology 9: 232-240.
- Wang L, Xie XQ (2014) Computational target fishing: what should chemogenomics researchers expect for the future of in silico drug design and discovery? Future medicinal chemistry 6(3): 247-249.
- Hopkins AL (2008) Network pharmacology: the next paradigm in drug discovery. Nature chemical biology 4(11): 682-690.
- Wang J, Gao L, Lee YM, Kalesh KA, Ong YS, et al. (2016) Target identification of natural and traditional medicines with quantitative chemical proteomics approaches. Pharmacology & therapeutics 162: 10-22.
- Lee YJ, Park BS, Park HR, Yu SB, Kang HM, et al. (2017) XIAP inhibitor embelin induces autophagic and apoptotic cell death in human oral squamous cell carcinoma cells. Environmental toxicology 32(11): 2371-2378.
- Avisetti DR, Amireddy N, Kalivendi SV (2019) The mitochondrial effects of embelin are independent of its MAP kinase regulation: Role of p53 in conferring selectivity towards cancer cells. Mitochondrion 46: 158-163.
- Avisetti DR, Babu KS, Kalivendi SV (2014) Activation of p38/JNK pathway is responsible for embelin induced apoptosis in lung cancer cells: transitional role of reactive oxygen species. PLoS One 9(1): e87050.
- Fu X, Pang X, Qi H, Chen S, Li Y, et al. (2016) XIAP inhibitor Embelin inhibits bladder cancer survival and invasion in vitro. Clinical & translational 18(3): 277-282.
- MK Parvez, M Tabish Rehman, P Alam, MS Al Dosari, SI Alqasoumi, et al. (2019) Plant-derived antiviral drugs as novel hepatitis B virus inhibitors: Cell culture and molecular docking study. Saudi pharmaceutical journal 27(3): 389-400.

 Research Article
Research Article