Abstract
Objective: Genome-wide analysis was used to screen differentially expressed genes after mild hypothermia treatment (MHT), and cluster analysis was performed to screen out and validate the target genes or pathways related to MHT in brain cortex of cardiac arrest rats after return of spontaneous circulation (ROSC).
Methods: Healthy adult male Wistar rats were induced ventricular fibrillation (VF) and CPR was performed after VF for 7 min. The rats were randomly divided into CPRT1 (36.5-37.5 ℃ ) and CPRT2 group (33.5-34.5 ℃). Three rats in each group were randomly sacrificed after ROSC for 2 hours, and cerebral cortex samples were taken to extract total RNA. RNA purification, amplification, and hybridization reaction with mouse whole gene chip to detect Differentially expressed genes which were further revalidated with realtime quantitative reverse transcriptase-polymerase chain reaction (Q-PCR) and Weston blot at 2h, 4h and 8h. Neurologic deficit score and TUNEL staining were assessed at 72 h after ROSC.
Results: MHT improved the NDS of rats and decreased the apoptotic cells in the cortex rat after ROSC for 72 hours. The genes associated with hypothermia and showing significant differential expression were p21, SFN, GADD45, BDNF, c-fos, HSP72, c-JUN, Nur77, CXCL2, CCL3, IL4 and TSLP. The MAPK signaling pathway, the cytokine-cytokine receptor interaction pathway and the P53 signaling pathway showed significant changes revalidated by qPCR and Weston blot.
Conclusion: Mild hypothermia exerts brain protections by affecting MAPK-associated inflammatory response, P53 apoptotic pathway and the cytokine receptor pathway after ROSC in rats.
Keywords: Mild Hypothermia;Genome- Wide Microarray;Cardiopulmonary Resuscitation; Rat; Brain Injury
Abbreviations: CPR: Cardiopulmonary Resuscitation; CA: Cardiac Arrest; ROSC: Return of Spontaneous Circulation; OHCA: Out of Hospital Cardiac Arrest; NDS: Neurological Deficit Score
Introduction
With the development of Cardiopulmonary Resuscitation (CPR) technology and the popularity of public CPR, the success rate of Cardiac Arrest (CA) patients with Return of Spontaneous Circulation (ROSC) has improved significantly, but less than 10% of patients are discharged with good neurologic outcome. Most patients die of hypoxic ischemic brain damage after CPR[1]. Mild hypothermia treatment can improve discharge survival rate and improve neurological function in Out of Hospital Cardiac Arrest(OHCA) patients[2, 3]. At present, the brain protection mechanism of TTM is still unclear.Biochip is a high-tech developed in the field of life sciences. The gene chip can detect the transcriptional changes of a large number of genes under different conditions, and can display gene expression levels reflecting characteristic tissue types, developmental stages, environmental condition responses, and genetic alterations, and become the preferred tool for highthroughput systems to investigate gene expression information of biological samples[4]. The effect of mild hypothermia on brain injury after ROSC is a complex and dynamic process, the traditional analytical methods cannot meet the need to analyze the expression of the entire genome in a single response. Therefore, in the current study, we plan to use the whole gene chip to analyze the effect of hypothermia on the gene expression in cortex of rat after ROSC, screen out differentially expressed genes, and elucidate its function, signaling pathway and network characteristics.
Materials and Methods
Animals
Male Wistar rats (aged 14–16 weeks and weighing 292.3– 402.5g) were provided by the Experimental Animal Center, Sun Yat-Sen University of Guangdong Province (Guangzhou, China). Ethical approval code: [2015] A-066. Animals were maintained on laboratory chow and housed in a specific pathogen-free room at a constant temperature (20–22 0C) with 10 h of light and 14 h of dark exposure. All animal studies were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University, and all experiments were performed in accordance with the Animal Research Reporting In Vivo Experiments guidelines on animal research[5].
Animal Model of Cardiac Arrest and Hypothermia
Male Wistar rats were randomized into 4 groups, normothermia control group (NT1, n=3), hypothermia control group (NT2, n=3), normothermia CPR group (CPRT1, n=18) and hypothermia CPR group (CPRT2, n=18). All the rats were anesthetized by intraperitoneal pentobarbital injection (30mg/kg; Sigma, USA). The procedures of induction of CA and CPR were done as our previously described[6,7] in CPRT1 and CPRT2 group but was not in NT1 and NT2 group. The rats in NT1 and CPRT1 group were kept in an incubator chamber to keep their esophageal temperature at 36.5-38.5℃. The rats in NT2 and CPRT2 group were placed ice cubes around the body to induce hypothermia after ROSC[8]. The esophageal temperatures were taken every 5 minutes and ice cubes were added or taken away to maintain their temperature between 33-35℃ for 12 hours and then gradually re-warmer at 0.5℃per hour.
Tissue Collection and RNA Extraction
Three of the rat’s cerebral cortex was harvested at 2 hours, 4 hours, and 8 hours after ROSC in CPRT1 and CPRT2 group, the brains were harvested. The cerebral cortices were snap-frozen in liquid nitrogen; after being complete frozen, they were stored in an airtight container at –800C.
mRNA Microarray
Total RNA was extracted from the cortex of 2 hours after ROSC in CPRT1, CPRT2 group and the animals in NT1, NT2 group using TRIZOL Reagent (Life technologies, USA) according to the manufacturer’s instruction. RNA was checked for integrity using Agilent Bioanalyzer 2100 (Agilent Technologies, USA) and quantified using Nanodrop ND-2000 (Thermo Fisher Scientific, USA). RNA was amplified and labeled using Low Input Quick Amp Labeling Kit, One-Color (Agilent Technologies, USA), followed by purification (RNeasy mini kit, Qiagen) and hybridization (Agilent Whole Rat Genome Microarray 4×44K, USA). Agilent Microarray Scanner was used for scanning the slides, and Agilent Feature Extraction Software was used for data extraction.
Gene Network Analysis
After data from mRNA microarrays were acquired, raw signals were log2 transformed. Differentially expressed mRNAs(DEMs) were screened for absolute values of fold change (FC) ≥ 2 and P-values < 0.05 (Student’s t-test). After that the preliminary candidate genes (P-value< 0.05, FC>2) underwent Hierarchical cluster analysisusing Gene Cluster 3.0 and Java Tree View 1.1.6 ,Go Gene Ontologyanalysis and KEGG enrichment analysis for functional analysis. Student’s t test and chi-square were employed to test for significance. P-value< 0.05 and FC>2 was used as the threshold for screening GO analysis and KEGG enrichment analysis.
Quantitative Reverse Transcription-PCR (qRT-PCR)
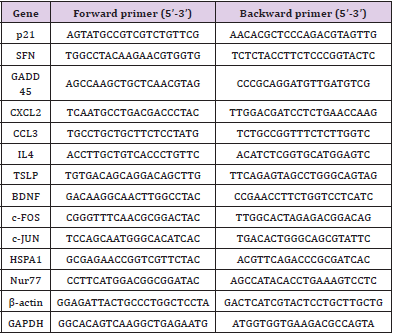
500ng of total RNA from 2,4 and 8 hours after ROSC in CPRT1, CPRT2 group were reversely transcribed using PrimeScriptTM RT Master Mix (Takara, Japan). Real-time PCR was performed using TG GreenTM Premix EX TaqTMⅡ(Takara, Japan) on LightCycler® 480 Ⅱ Real-time PCR Instrument (Roche Applied Science, Switzerland). β-actin was used as internal control. The primers used are listed in Supplementary Table 1.
Western Blotting
Total protein from 2, 4 and 8 hours after ROSC in CPRT1, CPRT2 group were extracted using RIPA (Millipore, 20188) with 1mM PMSF (CST, 8553) and protease inhibitor (Roche, complete ULTRA Tablets Mini EDTA-free). Protein concentrations were determined using a BCA Protein Assay Kit (Thermo Scientific, 23225). Protein extracts were heated to denature with Blue Loading Buffer (CST, 7722) according to manufacturer’s instruction. 20μg of each sample were loaded onto 12% SDS-PAGE gels and transferred to PDVF membranes. After blocking the membranes were incubated with primary antibodies against p21 (abcam, ab80633), NUR77 (abcam, ab109180), CXCL2 (abcam, ab25130), CCL3 (abcam, ab25128), IL4 (abcam, ab9811), TSLP (ProSci, 4025), SFN (abcam,ab193667), GADD45A (abcam, ab180768), BDNF (abcam, ab205067), c-Fos (abcam, ab209794), HSPA2 (abcam, ab108416), c-Jun (abcam, ab32137) and α-tubulin (santa cruz,sc-8035) respectively. HRP conjugate goat anti-rabbit IgG (Boster, BA1055) or goat anti-mouse IgG ( Boster, BA1051) were used as the second antibody. The blots were then visualized with an ECL kit (Millipore) in ChemiDoc Touch luminescent image analyser (BIO-RAD).
Neurological Deficit Score and TUNEL Staining
The other 12 rats in CPRT1 and CPRT2 group were induced VF and CPR, then were observed for 72 h after ROSC and their Neurological Deficit Score (NDS) were assessed from 0 (no observed neurological deficit) to 500 (death or brain death)[9] by two investigators who were blinded to the treatment each day. At 72 h postresuscitation, the rats were sacrificed. The cortex was collected for follow-up experiments.TUNEL staining was performed using the In-Situ Cell Death Detection Kit, POD (Roche Applied Sciences, Mannheim, Germany). Propidium Iodide staining was performed to visualize nuclei after TUNEL reaction. After these treatments, the cerebral cortex was analyzed by counting all TUNEL positive cells per 480×480 pixel in a magnification of 400-fold by light microscopy (OLYMPUS BX51, Tokyo, Japan). Electron microscopy (HITACHI CM10, Tokyo, Japan) was performed as well.
Statistical Analysis
The Statistical Program for Social Sciences (SPSS) 13.0 software (SPSS, Chicago, IL, USA) was used to perform all the statistical analyses. All data were expressed as the means ± SD or proportions where appropriate. For comparisons, unpaired t-tests were performed where appropriate between two groups. The correlation was calculated using Spearman’s correlation coefficient, which is the Pearson’s correlation coefficient of the indexed ranks of two data sets. P values of 0.05 (two-tailed) were considered statistically significant.
Results
DEMs Identified Between Groups
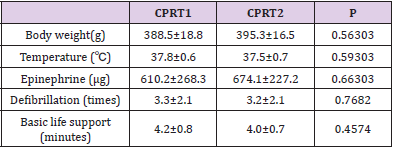
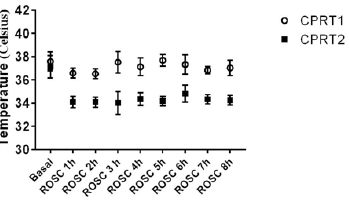
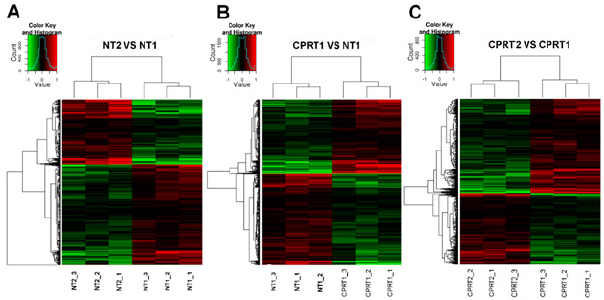
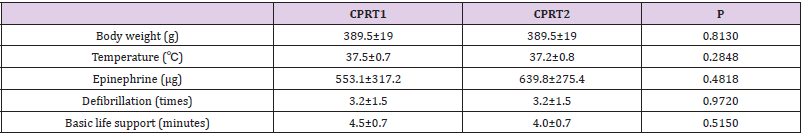
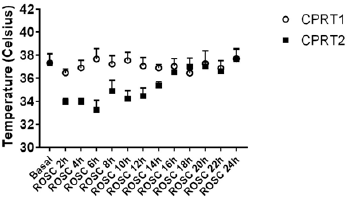
There were no differences of body weight, Temperature between CPRT1 and CPRT2 group at baseline. There were 13/18 rats in CPRT1 group and 12/18 rats in CPRT2 group gained the ROSC. The ROSC rate of rats, epinephrine dose, defibrillation times and base life support time were no difference between CPRT1 and CPRT2 group (supplementary Table 2). The temperature was controlled according to the protocol (supplementary Figure 1). Three rats in each time point at ROSC2h, 4h and 8h between two groups were scarified and the brain were harvested for further experiment. A total of 41012 mRNAs were detected in 4 groups. Hierarchical cluster analysis was performed to identify DEMs between groups. In NT2 group compared with NT1 group 138 DEGs were identified, with 96 DEMs (69.6%) up-regulated and 42 DEGs (30.4%) downregulated (Figure 1A). 227 DEGs were identified in CPRT1 samples compared with NT1 group, of which 150 DEMs (66.1%) were upregulated and 77 DEGs (33.9%) were down-regulated (Figure 1B). In CPRT2 group compared with CPRT1 group 1040 DEGs were identified, with 450 DEMs (43.3%) up-regulated and 590 DEGs (56.7%) down-regulated (Figure 1C).
Supplementary Table 2: CPR related parameters of rats for differentially expressed mRNAs analysis in CPRT1 and CPRT2 group.
Supplemenatry Figure 1: The body temperature control of rats for differentiallyexpressed mRNAs analysis in CPRT1 and CPRT2 group.
Figure 1: Hierarchical cluster analysis of DEMs between groups.
a) In NT2 group compared with NT1 group, 96 DEMs (69.6%) up-regulated and 42 DEGs (30.4%) down-regulated.
b) In CPRT1 group compared with NT1 group, of which 150 DEMs (66.1%) were up-regulated and 77 DEGs (33.9%) were down-regulated.
c) In CPRT2 group compared with CPRT1 group, of which 450 DEMs (43.3%) up-regulated and 590 DEGs (56.7%) downregulated.
Biological Function and Pathway Analysis of DEMs
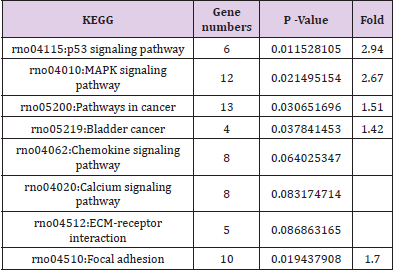
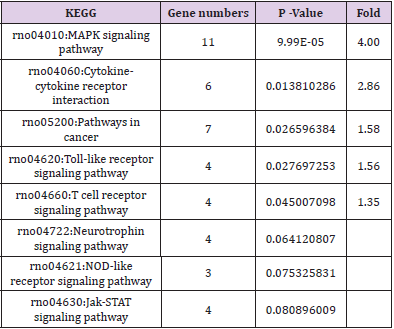
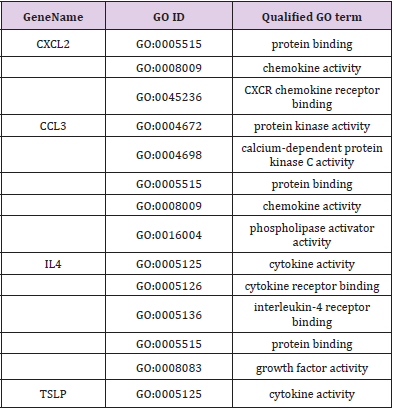
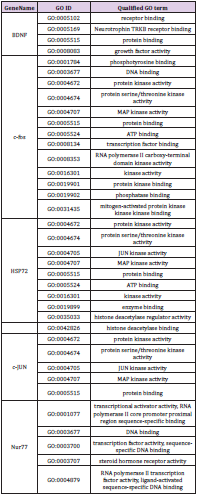
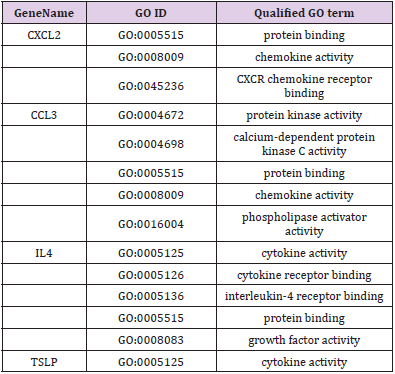
GO and KEGG analysis were employed to explain the biological function and find the enriched pathway of the DEMs in different groups. The differentially expressed genes of CPRT1 VS NT1 are mainly involved in protein binding, protein kinase activation, calcium-dependent protein kinase C activation, phospholipase activation, chemokine activation, chemokine receptor binding, and inflammatory factor receptors binding, inflammatory factor activation, etc indicating that CA-induced cerebral ischemiahypoxia- reperfusion injury can significantly increase intracranial inflammatory response. The differentially expressed genes in the CPRT2 VS CPRT1 group are mainly involved in protein binding, ATP binding, phosphatase binding, growth factor binding, neurotrophic receptor binding, encoding cyclins, cyclin binding, cyclin activation, RNA Polymerase II kinase activation, histone binding, core promoter binding, sequence-specific DNA binding, transcription factor binding, etc, which indicated that the effect of hypothermia on gene expression mainly focused on energy metabolism, brain neurotrophic, cell cycle regulation and apoptosis. The GO function of DEMs were shown in Supplementary Table 3-5.The signal pathway associated with CA/CPR brain injury in the CPRT1 VS NT1 group was p53 signaling pathway and the major differentially expressed genes were P21, 14-3-3-sigma (SFN) and GADD45 (Table 1). In CPRT2 VS CPRT1 group was the MAPK signaling pathway and the inflammatory factor receptor pathway. The main differentially expressed genes of MAPK signaling pathway are BDNF, c-fos, HSP72, c-JUN, GADD45 and Nur77; the main differentially expressed genes of inflammatory factor receptor pathway are CXCL2, CCL3, IL4 and TSLP (Table 2).
Supplementary Table 3: GO molecule function of differentially expressed mRNAs in the p53 signaling pathway in CPRT1 VS NT1 group.
Supplementary Table 4: GO molecule function of differentially expressed mRNAs in the MAPK signaling pathway in CPRT2 VS CPRT1 group.
Supplementary Table 5: GO molecule function of differentially expressed mRNAs in the cytokine-cytokine receptor interaction signaling pathway in CPRT2 VS CPRT1 group.
DEM Expression Tendencies following ROSC
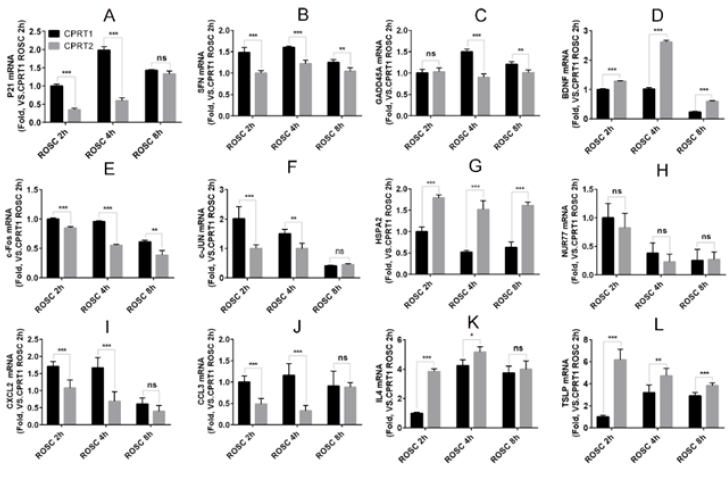
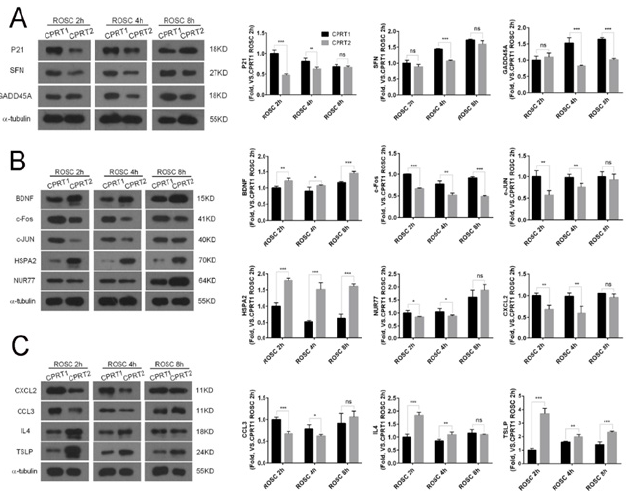
Hypothermia inhibits the expression of P21, SFN and GADD45A mRNA in P53 pathwaythe expression of P21 and SFN in ROSC 2h and 4h in CPRT2 group was significantly lower than that in CPRT1 group (Figure 2A& 2B), However, the effect on the expression of GADD45A was slightly later, the expression of GADD45A at ROSC at 4h and 8h in CPRT2 group was significantly lower than that in CPRT1 group (Figure 2C).Hypothermia promoted the expression of BDNF, HSPA2 at 2h, 4h, 8h after ROSC (Figure 2D& 2E), but inhibited the expression of c-Fos and c-JUN (Figure 2F & 2G).Hypothermia also had inconsistent effects on DEMs in the inflammatory factor receptor pathway, inhibiting the expression of CXCL2 and CCL3 at 2 h, 4 h after ROSC (Figure 2H, 2I & 2J), but promoting IL4 and TSLP expression (Figure 2K& 2L), especially for TSLP. We further observed the effect of Hypothermia on DEMs protein expression, CPRT2 group P21 (2h and 4h), SFN (4h) and GADD45A (4h and 8h) expression was lower than CPRT1 group in the p53 pathway (Figure 3A). The protein levels of BDNF (2h, 4h and 8h) and HSPA2 (2h, 4h and 8h) in CPRT2 group were significantly higher than those in CPRT1 group, while the expression of c-Fos (2h, 4h and 8h), c-JUN (2h and 4h) and NUR77 (2hand 4h) was lower than that of CPRT1 group in the MAPK pathway (Figure 3B). The protein levels of IL4 (2h and 4h) and TSLP (2h, 4h and 8h) in CPRT2 group were significantly higher than those in CPRT1 group, while the expression of CXCL2 (2h and 4h) and CCL3 (2hand 4h) was lower than that in CPRT1 group in inflammatory factor receptor pathway (Figure 3C).
Figure 2: The mRNA expressions tendency of DEMs after ROSC between CPRT1 and CPRT2 group.
A-C. The MHT decreased the mRNA levels of p21 (ROSC 2h, 4h), SFN (ROSC 2h, 4h and 8h) and GADD45 (ROSC 4h, 8h) after ROSC.
D-G. The MHT increased the mRNA levels of BDNF and HSP72 at 2h, 4h and 8h after ROSC, but decreased the mRNA levels of c-Fos (ROSC 2h, 4h and 8h) and c-JUN (ROSC 2h, 4h) after ROSC.
H-L. The MHT increased the mRNA levels of IL-4 (ROSC 2h, 4h) and TSLP (2h, 4h and 8h), decreased the mRNA levels of CXCL2 and CCL3 at 2h and 4h after ROSC, but no effects on NUR77 expression.
Figure 3: TThe protein expressions tendency of DEMs after ROSC between CPRT1 and CPRT2 group.
a) The MHT decreased the protein levels of p21 (ROSC 2h, 4h) SFN (ROSC 4h) and GADD45 (ROSC 4h, 8h) after ROSC.
b) The MHT increased the protein levels of BDNF and HSP72 at 2h, 4h and 8h after ROSC, but decreased the protein levels of c-Fos (ROSC 2h, 4h and 8h) and c-JUN (ROSC 2h, 4h) after ROSC.
c) The MHT increased the protein levels of IL-4 (ROSC 2h, 4h) and TSLP (2h, 4h and 8h), decreased the protein levels of CXCL2 and CCL3 at 2h and 4h after ROSC.
Hypothermia Treatment Improved Neurologic Deficit Scores and Decreased Neuronal Apoptosis following ROSC
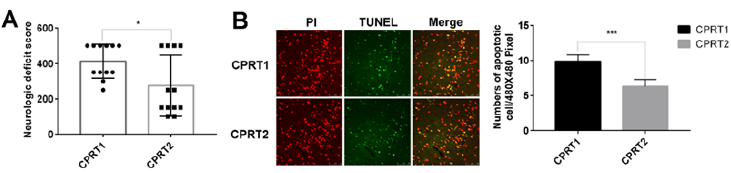
The induction of Ventricular Fibrillation (VF) caused serious injury to the brain in rats, resulting in the death of 6/12 rats within 72 h in the CPRT1 group, whereas 8/12 rats survived to 72 h in the CPRT2 group. The ROSC rate of rats, epinephrine dose, defibrillation times and base life support time were no difference between CPRT1 and CPRT2 group (Supplementary Table 6). The temperature was controlled according to the protocol (Supplementary Figure 2).The NDS in the CPRT1 group were 425 ± 87, which were significantly higher than those (288 ± 48) in the CPRT2 group (P=0.0338) (Figure 4A). TUNEL-positive nuclei were observed in the cortex of animals in both CPRT1 and CPRT2 groups. The number of apoptotic neurons was 9.89 ± 0.9 in the CPRT1 group, higher than the 6.3 ± 1.0 apoptotic neurons observed per 480 × 480 pixel in the 40× field in the CPRT2 group (P< 0.001) (Figure 4B).
Supplementary Table 6: CPR related parameters of rats for neurologic function assessment in CPRT1 and CPRT2 group.
Supplemenatry Figure 1: The body temperature control of rats for differentiallyexpressed mRNAs analysis in CPRT1 and CPRT2 group.
Supplemenatry Figure 2: The body temperature control of rats for differentiallyexpressed mRNAs analysis in CPRT1 and CPRT2 group.
Figure 4: Hypothermia Treatment Improved Neurologic Deficit Score and Decreased Neuronal Apoptosis after ROSC
a) Hypothermia Treatment Improved Neurologic Deficit Score of rats after ROSC for 72 hours.
b) Hypothermia Decreased Neuronal Apoptotic cells in the cortex of rats after ROSC for 72 hours. (scale bar =50 μm)
Discussion
For the first time, we used genome-wide microarray to study the effects of MHT on gene expression profile changes after cardiopulmonary resuscitation and explored the mechanism of MHT on brain protection from single gene changes to gene related pathways or network. The rat CA/CPR model was used in this study, which is closer to the evolution of cerebral ischemia-reperfusion injury after CA/CPR in clinical practice.A total of 41012 mRNAs were detected and hierarchical cluster analysis was performed to identify DEMs between groups. GO and KEGG analysis were employed to explain the biological function and find the enriched pathway of the DEMs in different groups. This study found that MHT improved neurological function of rats after CA/CPR by affecting p53 signaling pathway, MAPK signaling pathway and inflammatory factor receptor pathway. The major DEMs are P21, 14-3-3-sigma (SFN) and GADD45 in P53 pathway. The main DEMs of MAPK signaling pathway are BDNF, c-fos, HSP72, c-JUN and Nur77. The main DEMs of inflammatory factor receptor pathway are CXCL2, CCL3, IL4 and TSLP. P53 and its downstream genes play an essential role in cerebral ischemic cell apoptosis[10]. Downstream genes of p53 have two functions, the regulation of the cell cycle and the regulation of cell apoptosis[11]. After ROSC, the expression of p21, SFN (14-3-3-σ), GADD45 is up-regulated in the cerebral cortex of rats.
These genes are involved in inflammatory responses, cell cycle arrest and apoptosis[12,13], and MHT can inhibit the expression of these genes.The MAPK family is involved in cerebral ischemia. It is well documented that ERK1/2 modulates neuronal survival and apoptotic cell death. Activation of this complex results in phosphorylation of many cytoplasm and membrane proteins[14]. Previous research has also reported that activation of JNK and p38 MAPK is mediated in neuronal apoptosis, infracted volume, and neurological deficits in ischemic stroke[15]. The present study shows that MAPK signaling pathways were regulated differently by MHT, which increases expressions of BDNF and HSP72, but inhibits the expression of c-fos, c-JUN and Nur77. BDNF is a member of the family of neurotrophies in the central neural system. As an attractive target gene of CREB[16], the mature BDNF plays a vital role in antiinflammation, anti-neurotoxicity, promoting neuronal survival and regeneration following ischemic brain injury[17]. HSP72, the major inducible member of the heat shock protein 70 family, has been found protecting cells from certain apoptotic stimuli such as oxidative stress, hypoxia and inflammation[18], could decreased the activation of JNK3, c-Jun and caspase-3 induced by cerebral I/R[19].
The signals of c-fos, c-jun, and nur77 were induced with different degree of intensity by hypoxia and were reduced significantly by naloxone have protections on PC12 cells survival after hypoxia[20]. Expression of c-Jun and caspase-2 is associated with neuronal cell apoptosis in the retinal ganglion cell layer[21]. The up-regulation of Nur77 mediated neuron apoptosis and mitochondrial injury via aberrant mitochondrial fragmentation in a manner dependent on the Wnt/β-catenin/INF2 pathway, while ablation of Nur77 resulted in a reduction in the infarction area, decreased neuronal apoptosis and attenuated mitochondrial injury[22]. MHT selectively acts on different components of the MAPK pathway, increasing the expression of components beneficial to neuronal survival, reducing the expression of components involved in apoptosis. The mechanism of differential effects of MHT on MAPK is not clear, it may be related to the spatial discrepancy expression of these gene[23].Inflammatory processes play a fundamental role into brain ischemia-reperfusion injury[24]. in the present study, we found that the DEMs of inflammatory factor receptor pathway are CXCL2, CCL3, IL4 and TSLP, the MHT increases the expression of IL4 and TSLP but decreases the expression of CXCL2 and CCL3. TSLP is also expressed in the Central Nervous System (CNS) where it is produced by choroid plexus epithelial cells and astrocytes in the spinal cord[25].
Study showed that TSLP involved in the pathogenesis of ischemic stroke[26] contributed to angiogenesis which is a key neuro-restorative event in response to ischemia. The cytokine IL-4 improves long-term neurological outcomes after stroke, perhaps through M2 phenotype induction in microglia/macrophages, immunomodulation with IL-4 is a promising approach to promote long-term functional recovery after stroke[27]. The expression of CXCR2 increased in the ischemic brain correlated with increased leukocyte accumulation in the ischemic brain after focal stroke. Cytokine-induced neutrophil chemoattractant-1 was the major chemokine involved in neutrophil recruitment to the brain[28]. CCL2, CCL3 and CCL5 recruit monocytes and T cells via the chemokine receptors CCR1, CCR3, and CCR5. Increased levels expression and production of CCL3 have been described in experimental brain stroke[29].There are some limitations in the current study, the first one is the only one section of the genomewide analysis was observed, which could not fully reflect the effect of MHT on cerebral cortex gene changes after ROSC. However, when DEMs were verified at 2 hours, 4h and 8h after ROSC, its tendency was consistent with the genome-wide analysis anticipated. The second one is that the components of cerebral cortex cells are complex, and the selected DEMs can only reflect the overall changes in the entire cerebral cortex. Whether these changes are caused by neuronal cells, glial cells, and infiltrating inflammatory cells required further study to confirm, but it gave hints for future research.
Conclusion
MAPK-associated inflammatory pathway, the P53 apoptotic pathway, and the cytokine receptor pathway are associated with brain damage after cardiopulmonary resuscitation. MHT exerts brain protection by affecting the inflammatory response, apoptosis, and cytokine receptor-mediated damage pathway after ROSC.
Acknowledgment
This study was supported by funding from the Natural Science Foundation of Guangdong province (2020A1515010120), National Nature Science Foundation of China (81272021, 81571867, 81901931) and Shenzhen Science and Technology Plan Project (JCYJ20160608142215491). The funders had no role in the study design, data collection, analysis, decision to publish, or the preparation of the manuscript.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Cronberg T, Brizzi M, Liedholm LJ (2013) Neurological prognostication after cardiac arrest--recommendations from the Swedish Resuscitation Council. Resuscitation 84(7): 867-872.
- Michael Holzer (2002) Mild therapeutic hypothermia to improve the neurologic outcome after cardiac arrest. N Engl J Med 346(8): 549-556.
- Bernard SA, Gray TW, Buist MD (2002) Treatment of comatose survivors of out-of-hospital cardiac arrest with induced hypothermia. N Engl J Med 346(8): 557-563.
- Zhu T (2003) Global analysis of gene expression using Gene Chip microarrays. Curr Opin Plant Biol 6(5): 418-425.
- Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG (2011) Animal research: reporting in vivo experiments--the ARRIVE guidelines. J Cereb Blood Flow Metab 31(4): 991-993.
- Wei H, Yin M, Lu Y (2019) Mild hypothermia improves neurological outcome in mice after cardiopulmonary resuscitation through Silent Information Regulator 1-actviated autophagy. Cell Death Discov. 5: 129.
- Lu Y, Zeng X, Jing X (2019) Pre-arrest hypothermia improved cardiac function of rats by ameliorating the myocardial mitochondrial injury after cardiac arrest. Exp Biol Med (Maywood) 244(14): 1186-1192.
- Chun Lin H, Jie W, Xiao Xing L (2011) Effects of therapeutic hypothermia on coagulopathy and microcirculation after cardiopulmonary resuscitation in rabbits. Am J Emerg Med 29(9): 1103-1110.
- Hendrickx HH, Rao GR, Safar P, Gisvold SE (1984) Asphyxia, cardiac arrest, and resuscitation in rats. I. Short term recovery. Resuscitation 12(2): 97-116.
- Tu YF, Lu PJ, Huang CC, Ho CJ, Chou YP (2012) Moderate dietary restriction reduces p53-mediated neurovascular damage and microglia activation after hypoxic ischemia in neonatal brain. Stroke 43(2): 491-498.
- Guo YB, Bao XJ, Xu SB, Zhang XD, Liu HY (2015) Honokiol induces cell cycle arrest and apoptosis via p53 activation in H4 human neuroglioma cells. Int J Clin Exp Med 8(5): 7168-7175.
- LaBaer J, Garrett MD, Stevenson LF (1997) New functional activities for the p21 family of CDK inhibitors. Genes Dev 11(7): 847-862.
- Grasso D, Garcia MN, Hamidi T (2014) Genetic inactivation of the pancreatitis-inducible gene Nupr1 impairs PanIN formation by modulating Kras(G12D)-induced senescence. Cell Death Differ 21(10): 1633-1641.
- Zhao TT, Kim KS, Shin KS (2017) Gypenosides ameliorate memory deficits in MPTP-lesioned mouse model of Parkinson's disease treated with L-DOPA. BMC Complement Altern Med 17(1): 449.
- Wu X, Li L, Zhang L (2015) Inhibition of thioredoxin-1 with siRNA exacerbates apoptosis by activating the ASK1-JNK/p38 pathway in brain of a stroke model rats. Brain Res 1599: 20-31.
- Terasaki Y, Sasaki T, Yagita Y (2010) Activation of NR2A receptors induces ischemic tolerance through CREB signaling. J Cereb Blood Flow Metab 30(8): 1441-1449.
- Han BH, Holtzman DM (2000) BDNF protects the neonatal brain from hypoxic-ischemic injury in vivo via the ERK pathway. J Neurosci 20(15): 5775-5781.
- Park HS, Cho SG, Kim CK (2002) Heat shock protein hsp72 is a negative regulator of apoptosis signal-regulating kinase 1. Mol Cell Biol 22(22): 7721-7730.
- Qi D, Liu H, Niu J (2012) Heat shock protein 72 inhibits c-Jun N-terminal kinase 3 signaling pathway via Akt1 during cerebral ischemia. J Neurol Sci 317(1-2): 123-129.
- Huang HM, Yu JY, Ou HC, Jeng KC (2008) Effect of naloxone on the induction of immediately early genes following oxygen- and glucose-deprivation in PC12 cells. Neurosci Lett 438(2): 252-256.
- Kurokawa T, Katai N, Shibuki H (1999) BDNF diminishes caspase-2 but not c-Jun immunoreactivity of neurons in retinal ganglion cell layer after transient ischemia. Invest Ophthalmol Vis Sci 40(12): 3006-3011.
- Zhao H, Pan W, Chen L, Luo Y, Xu R (2018) Nur77 promotes cerebral ischemia-reperfusion injury via activating INF2-mediated mitochondrial fragmentation. J Mol Histol 49(6): 599-613.
- Kinoshita Y, Ueyama T, Senba E, Terada T, Nakai K (2001) Expression of c-fos, heat shock protein 70, neurotrophins, and cyclooxygenase-2 mRNA in response to focal cerebral ischemia/reperfusion in rats and their modification by magnesium sulfate. J Neurotrauma 18(4): 435-445.
- Rodríguez Yáñez M, Castillo J (2008) Role of inflammatory markers in brain ischemia. Curr Opin Neurol 21(3): 353-357.
- Kitic M, Wimmer I, Adzemovic M (2014) Thymic stromal lymphopoietin is expressed in the intact central nervous system and upregulated in the myelin-degenerative central nervous system. Glia 62(7): 1066-1074.
- Yu X, Peng Y, Liang H (2018) TSLP/TSLPR promote angiogenesis following ischemic stroke via activation of the PI3K/AKT pathway. Mol Med Rep 17(2): 3411-3417.
- Liu X, Liu J, Zhao S (2016) Interleukin-4 Is Essential for Microglia/Macrophage M2 Polarization and Long-Term Recovery After Cerebral Ischemia. Stroke 47(2): 498-504.
- Wang X, Li X, Schmidt DB (2000) Identification and molecular characterization of rat CXCR3: receptor expression and interferon-inducible protein-10 binding are increased in focal stroke. Mol Pharmacol 57(6): 1190-1198.
- Wolinski P, Glabinski A (2013) Chemokines and neurodegeneration in the early stage of experimental ischemic stroke. Mediators Inflamm 2013(8): 727189.

 Research Article
Research Article