ABSTRACT
Background: Definition extraction and/or generation is an important task of information extraction and has proven useful in many applications such as intelligent question and answering systems, especially in this big data era. Most of the current researches about definition extraction focused on lexico-syntactic patterns or word lattices to identify definitional sentences, which usually suffered from poor performance due to the noisy and variable syntactic structures and word lattices in the real-world documents and texts.
Methods: This paper presents a straightforward approach to generate definition for medical terminology using the definitional relations from well-developed Medical Knowledge Organization System (MKOS), which will largely improve the accuracy and reliability of the results as the depended relations have already been reviewed and verified by the editors and domain experts of MKOS. Besides, two approaches of definition visualization were theoretically adopted and practically implemented to help the user intuitively understand the inherent nature of the generated definitions, which is firstly named as “definition visualization”.
Results: To evaluate and verify the performance of the proposed methods, a big number of testing data from well-known MKOS were collected to conduct the experiments. Experimental results verify our approaches by showing exactly suitable statistical values for human reading and ordinary file memory, as well as promising precision and feedback from domain experts.
Conclusion: The proposed approaches are able to generate precise definition based on the existing MKOS and will also innovatively convey the inherent nature and meaning of the generated definition in terms of two graphics diagrams.
Keywords: Definition Generation; Definition Visualization; Definitional Relationship; Medical Knowledge Organization System; Intelligent Question and Answering
Introduction
The age of big data speeds up the development of intelligent question and answering systems, especially in the biomedical field where a great deal of health data emerges all the time. While people search the basic meaning and intension of a medical terminology (also called term), textual definition will become a fundamental data source, thus various kinds of medical knowledge bases and domain glossaries immediately come to mind for consultation purposes. Unfortunately, these systems and glossaries will probably be of no use mainly due to the low precision of results. Therefore, it is quite natural to explore methods to extract or generate accurate definition for medical terminology automatically. Fortunately, these algorithms are quite promising and achieve a lot of importance and applications. Typical usage is the intelligent question and answering systems to reply a query such as “what is the Alzheimer Disease” [1-7].
Generally speaking, the current research about term definition extraction or generation lies in the observation that, some textual contexts in which a term occurs provide a formal explanation of the term of interest and may be used to generate definition with proper processing and further organization. These works have been adopted in several languages, for instance English [8], Chinese [9], German [10], Portuguese [11] and so on. And the particular implementation has been done either semi-automatically or fully automatically. In the former group, patterns are usually pregenerated from simple sentences of words, such as X refers to Y, X is defined as Y, or X is a Y. Most of the recent work’s attribute to the latter group [12-16]. Unlike regular expression-based hard matching patterns, Cui et al. [12,13] showed that soft patterns could model language variations probabilistically to extract definitional sentences and they later presented a new approach [14] by using probabilistic lexico-semantic, soft matching patterns based on bigrams and Profile Hidden Markov Model (PHMM) to identify definition sentences; Their experimental results showed that both models outperformed hard matching patterns by allowing a partial matching, while PHMM was more capable of dealing with language variations. Borg et al. explored the genetic programming to learn the typical linguistic forms of definitions and proposed a genetic algorithm to learn the relative importance of these forms [15], which could be able to learn similar rules derived by a human linguistic expert and rank candidate definitions in an order of confidence. Navigli et al. proposed a generalization, Word-Class Lattices learned from Wikipedia dataset to model textual definitions [8], which compared favorably to other algorithms in the terms of no parameter tuning and being capable for quite complex task (as in real-world documents).
Recently, Anke et al. provided a supervised approach and only used the syntactic features derived from dependency relations [16], where the problem was modeled as a classification task and each sentence had to be classified as being or not definitional. They got promising result by comparing with one well-known supervised and one unsupervised method. However, in the realworld documents and texts, as the definitional sentences often occur in highly variable syntactic structures and the definitional patterns are inherently very noisy, this kind of methods eventually becomes not aggressive in terms of low recall and precision. In contrast with the above algorithms, we present a straightforward approach to generate definition from the definitional relations in the well-developed MKOS, which will largely improve the accuracy and reliability of the results because the depended relations have already been reviewed and verified by the editors of MKOS, as well as the domain experts involved in the development of the MKOS. Besides, in order to help the user better understand the inherent nature of the generated definitions, we then implement two approaches of scientific and information visualization to intuitively convey the definition of terminology in terms of graphic diagrams, which is firstly regarded as “definition visualization”.
Theory and Approach for Medical Definition Generation
Brief of MKOS
MKOS is a dictionary or glossary consists of a large number of concepts in a domain, their various names and the relations among them, such as a thesaurus, ontology and so on. In biomedicine and healthcare area, Systematized Nomenclature of Medicine - Clinical Terms (SNOMED CT) is considered to be the most comprehensive and precise clinical health terminology product in the world, as well as a systematically organized computer interoperable collection consists of medical terms providing codes, terms, synonyms and relations used in clinical documentation and reporting [17]. The primary purpose of SNOMED CT aims to facilitate the accurate recording and sharing of clinically related health information and the semantic interoperability of health records. Now, SNOMED CT covers most of the core general clinically relevant concepts and terminologies, including body structures, diseases, clinical finding, operation, organisms and drugs. In 1999, SNOMED CT was established by the combing, expansion and reconstruction of two well-developed knowledge organization systems: one is SNOMED Reference Terminology (SNOMED RT) from the College of American Pathologists (CAP) and the other is Clinical Terms Version 3 (CTV3) owned by the National Health Service of the United Kingdom (NHS). Currently, SNOMED CT is owned and distributed around the world by the SNOMED Standards Development Organization (SDO) in the formal creation of the International Health Terminology SDO (IHTSDO).
Being a knowledge organization system, SNOMED CT is a core clinical healthcare terminology that contains concepts with unique meanings and formal logic based definitions organized into hierarchies. SNOMED CT content is organized with three kinds of components: concepts (from clinical meanings), descriptions (synonyms and other terms referred to concepts) and relationships (connecting one concept with other related concepts). Inside the 2015 version, more than 300,000 unique SNOMED CT concepts are grouped into 19 branches (top levels) of the hierarchies and organized from the general to the more detailed: Clinical finding, Procedure, Situation with explicit context, Observable entity, Body structure, Organism, Substance, Pharmaceutical / biologic product, Specimen, Special concept, Physical object, Physical force, Event, Environments and geographical locations, Social context, Staging and scales, Qualifier value, Record artefact and SNOMED CT Model Component. Besides, there are about 100 kinds of relationship between the concepts from SNOMED CT, such as is_a, has_finding_ site, has _associated_morphology, due_to, has_pathological_process, has_procedure_site, has_specimen and so on.
Differentiating Definitional from Non-Definitional Relationships in SNOMED CT
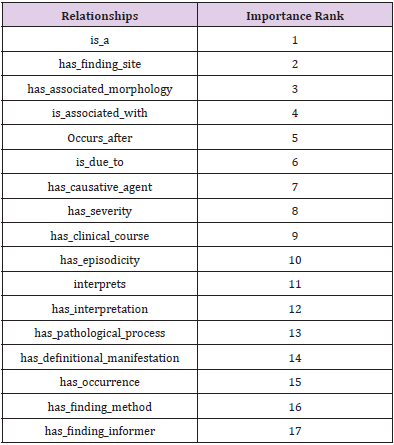
Among the large scale concepts and relationships, most of the relationships are used to define and represent the meaning of SNOMED CT concepts in these 9 branches of hierarchies: Clinical finding concepts, Procedure concepts, Evaluation procedure concepts, Specimen concepts, Body structure concepts, Pharmaceutical/biologic product concepts, Situation with explicit context concepts, Event concepts and Physical object concepts. Take the clinical finding concept as an example, the set of definitional relationships are shown in Table 1, sorted by the importance rank in a descending order. About the detailed definitional relationships and the brief description of their meanings used to define clinical finding concepts and other concepts, please refer to the Start Guide documentation of SNOMEDCT from its homepage [17].
Generating Term Definition from Definitional Relationships in SNOMED CT
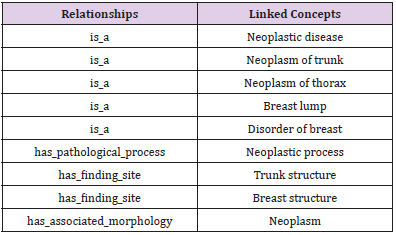
With the definitional relationship from SNOMED CT, we could generate the definition for a specific concept or terminology with further organization. For example, there are 9 definitional relationships about breast neoplasm (a kind of clinical finding) in SNOMED CT, see Table 2. With these definitional relationships and the relevant concepts names linked to clinical finding concepts, we could generate definitions for these terminologies following the rules below:
a) Firstly, we list the terminology of interest, namely the preferred term of one concept (PT), as a concept is usually expressed by a set of synonyms in MKOS
b) Specify the direct parent of hierarchy, which gives the inherent nature of the terminology of interest and is usually written as is_a;
c) Indicate other definitional relationships as well as the correlated concepts, ordered by the importance of the relationships; for the concepts from SNOMEDCT, this kind of importance has already been numbered and recorded in Start Guide documentation (as shown in Table 1); for other MKOS, it is suggested to get the importance from source documents or domain experts.
d) In case of many individuals (e.g., linked concepts) among one relationship type, list all of them in alphabetical order and take a comma to make a distinction between each other.
e) Separate each kind of relationships with a semicolon and end the definition sentence with a point.
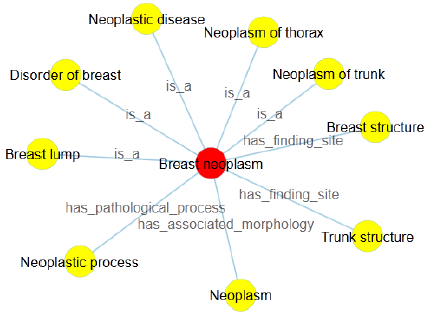
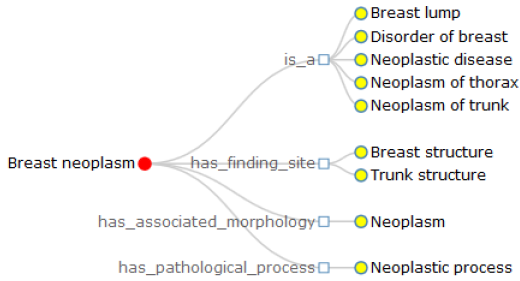
According to the above rules, the definition of breast neoplasm in Table 2 may be written as: Breast neoplasm is a Breast lump, Disorder of breast, Neoplastic disease, Neoplasm of trunk, Neoplasm of thorax; has finding site Breast structure, Trunk structure; has associated morphology Neoplasm; has pathological process neoplastic process. It is clear that this kind of definition holds valuable information, as it defines characteristics to distinguish meaning of one terminology from other similar concepts. However, it is easy to point out two grammar errors from these definitional sentences. One is that the sentences 2-5 are lack of the subjects, the other is the linked concepts related to breast neoplasm initial in capitals (except all capital words, such as AIDS). We then add two supplementary rules:
a) Start the sentences with ‘It’ except the first one, which is used to refer to the terminology of interest
b) Change the first capital letter of the linked concept into small one, except all capital words.
Therefore, the definition of breast neoplasm will be further modified and optimized as: Breast neoplasm is a breast lump, disorder of breast, neoplastic disease, neoplasm of trunk, neoplasm of thorax; It has finding site breast structure, trunk structure; It has associated morphology neoplasm; It has pathological process neoplastic process.
Theory and Approaches for Medical Definition Visualization
Scientific and information visualization is a branch of computer graphics used to improve understanding of the data or information being presented by means of images, which focuses on the creation of ways for presenting abstract data and information in intuitive ways. So far, there are many programing techniques used to realize visualization, such as JavaScript, Flash, Silverlight, Java Applet and so on. Among them, JavaScript is the best choice for dynamically visualizing data or information on the web page, as it shows a faster speed of image plotting compared to Silverlight and Java Applet and also works independently without the plug-in components (but Flash needs). D3.js is one of the famous JavaScript libraries for manipulating documents based on data and information, initially developed by the Computer Science Department of Stanford University [18,19]. D3 means Data-Driven Document, which firstly binds arbitrary data and information to a Document Object Model (DOM) and then applies data-driven transformations to the document. One of D3’s extremely advantages is fast, as well as supporting large datasets and dynamic behaviors for interaction and animation owing to minimal overhead. Moreover, D3 emphasizes on web standards and gives user the full capabilities of modern browsers. Up to now, D3 has already supported a large number of visualization applications with a variety of plotting charts and diagrams, for instance, HTML table from an array of numbers, interactive SVG bar chart with smooth transitions and interactions, force-directed graph showing character co-occurrence and so on. And D3 has already been applied for visualization of KOS dynamically and interactively [20].
In this research, in order to intuitively convey the definition of terminology, we implement the node-link and the right-oriented tree diagrams based on the layout of D3.js (D3.layout). Here, we presume that the node-link tree layout highlights the relationships between the root and the leaf nodes with a ragged appearance supported by the Cartesian orientations, while the right-oriented tree diagram clearly shows the class hierarchy and aligns the leaf nodes of the tree on the right edge. Inside the visualization image, the terminology of interest is the root node, the linked concepts become the leaf nodes, joined by various relationship types. Further with the definition of breast neoplasm, we get two approaches of definition visualization, Figures 1 & 2, with different colors to make a distinction between the root node and linked terms. Specifically, breast neoplasm is the root node in the visualization diagrams with red color, while other concepts are drawn in yellow color, connected by the corresponding relationship types. Note that in Figure 2, we group the relationships into several types and draw them with the rectangle shapes, with the aims to highlight the relations as well as the leaf nodes. We believe these two kinds of definition visualization will take advantage of the human eye’s broad bandwidth pathway into the mind and help users to see, explore and understand the inherent nature of the interested terminology at once.
Experimental Testing and Discussion
Experimental Setup
We conducted experiments on the dataset of the clinical finding branch from SNOMED CT, which contains a total number of 97,646 about clinical finding concepts and 2,338,582 relationships, grouped into 224 types of relationships. As shown above, there are 17 definitional types of relationships (Table I), involving 717,234 definitional relationships. The reason for using the hierarchy branch about clinical finding is that it primarily consists of the disease names (e.g., pneumonia) as well as the common signs and symptoms (e.g., fever, cough) known to mankind. Thus, clinical finding keeps the most importance to the public as it tightly closes to our healthcare. Besides, clinical finding is regarded as an application and reuses other branches of concepts, such as body structure, substance (e.g., drugs), procedure and so on. In other words, it is not possible to represent or describe the definitional content of clinical finding without explicitly or implicitly referring to anatomical concepts or chemical entities. For example, pneumonia must take for granted the existence of the lung structure and also the fully-defined diagnosis.
Measures
To assess the performance of our algorithm, we will calculate the following statistical measures:
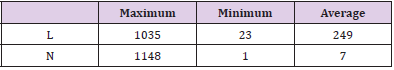
a) Length (L): The number of characters in each definition sentence. A definition is too long to read for most people, while it is almost useless if it is too short. Therefore, the maximum, minimum and average L will be calculated one by one.
b) Number (N): The number of the definitional relationships used to generate the definition of each terminology. It is quite obvious that a definition will contain a large amount of information if it is generated from lots of definitional relationships. However, there might be an implicit correlation between L and N, as a big N always leads to a large L. Similarly, the maximum, minimum and average N will be computed separately.
c) Accuracy (A): The number of correct definitions accepted by the domain experts over the total number of definitional sentences extracted for human evaluation and assessment.
Results and Discussion
Using the proposed approaches for definition generation and visualization, we get 100,332 definitions of terminologies about clinical finding. In Table 3, we report the results of statistical measures. The results show that the maximum length of the definitions contains 1035 characters, while the minimum length is 23. And the average length is 249, which will be exactly suitable for human reading as well as ordinary file memory (e.g., 255 characters for common text such as Microsoft Excel and Access). Besides, from the dimension of the definitional relationships each terminology owns, the maximum number is 1148, while minimum becomes 1. And the average is 7, means that most of the definitional relationships used to generate definitions contains considerable information to fully define the terminologies of interest.
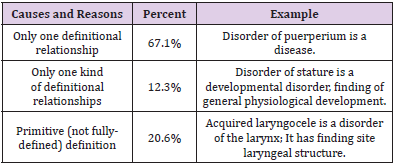
We then randomly extract 1000 definitional sentences for accuracy measures. With the help of the definition visualization (especially in the case that there are a large number of definitional relationships), two domain experts on biomedical informatics read these definitions and finally accepted 938, which means the accuracy is 93.8%. With regard to the unaccepted definitions by experts, we then manually analyze these 62 sentences and identify the possible causes and reasons in detail. The results are shown in Table 4. Two explicit and important causes (79.4%) are that there is only one definitional relationship or one type of definitional relationships about the terminology in SNOMED CT, which leads to the generated definitional sentence having insufficient information. The other reason (20.6%) is due to the primitive (not fully-defined) definition, as it is inadequate to uniquely distinguish its meaning from other similar concepts. However, the unaccepted definition could be further used as annotation or comment, which might make a reference to a specific explanation of terminology.
At the last part of this section, we would like to discuss the performance of the proposed definition visualization. From the feedback of the auditing experts as well as our practical experience, the proposed approaches of definition visualization will be of great assistance to better understand the definitional sentence and the inherent nature of the terminology of interest under two typical cases:
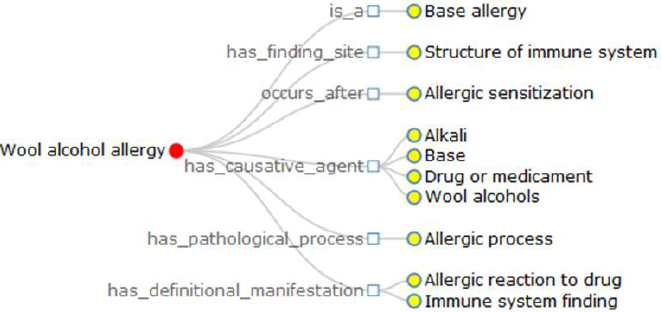
a) There is a big number of the definitional relationships used to generate the definition of terminology. For instance, we get the definition of wool alcohol allergy as: Wool alcohol allergy is a base allergy; It has finding site structure of immune system; It occurs after allergic sensitization; It has causative agent alkali, base, drug or medicament, wool alcohols; It has pathological process allergic process; It has definitional manifestation allergic reaction to drug, immune system finding. Totally, this sentence contains 10 definitional relationships grouped into 6 kinds. And a human being is quite difficult to quickly catch the key points, but the visualization does (Figure 3).
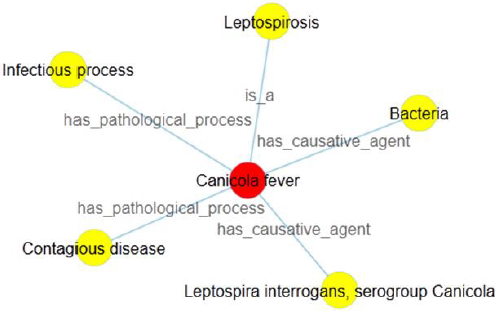
b) There is a comma inside the related concept. According to the former rule 4 of the definition generation, in case of many individuals (linked concepts) among one relationship type, we add a comma into the definition sentence and make a distinction between each other. By coincidence, the expression form of linked concept also consists of a comma. Then it is not easy for the user to exactly segment sentence and make pause without the aid of definition visualization. Given an example of canicola fever, we generate its definition sentence as: Canicola fever is a leptospirosis; It has causative agent bacteria, leptospira interrogans, serogroup canicola; It has pathological process contagious disease, infectious process. Here, Figure 4 intuitively tells us that “bacteria” and “leptospira interrogans, serogroup canicola” are the causative agents of canicola fever, but not “bacteria”, “leptospira interrogans” and “serogroup canicola”.
Conclusion
In this paper, we have presented a straightforward approach to generate the definition of medical terminology based on the definitional relationships about that terminology from welldeveloped MKOS. We also implemented two approaches of definition visualization and intuitively conveyed the definition of terminology in terms of graphic diagrams, which could help the user better understand the inherent nature of the interested terminology. To evaluate and verify the performance of the proposed methods, we have conducted the experiments using a large number of testing data from SNOMED CT, which is the most comprehensive and precise clinical health terminology product in the world. Testing results showed a promising performance in terms of statistical measures as well as practical feedbacks from domain experts. While we introduce and evaluate the presented algorithms only on data from MKOS in the biomedicine and healthcare area, we believe that our basic principles for definition generation and visualization are generic and can be extended to other areas where there are a great number of definitional relationships of terminologies, followed by the applications in intelligent question and answering widely used in this AI powered big data era.
Financial Support
This research is based on work supported (in part) by National Social Science Fund of China (No. 20BTQ062). Besides, the authors sincerely appreciate the editors and the anonymous reviewers who offer valuable suggestion and comments to help improve the quality of the manuscript.
Conflicts of Interest
There are no conflicts of interest.
References
- Demner Fushman D, Mrabet Y, Ben Abacha A (2020) Consumer health information and question answering: helping consumers find answers to their health-related information needs. J Am Med Inform Assoc 27(2): 194-201.
- Ren J, Liu N, Wu X (2020) Clinical questionnaire filling based on question answering framework. Int J Med Inform 141: 104225.
- Sarrouti M, Ouatik El Alaoui S (2017) A Machine Learning-based Method for Question Type Classification in Biomedical Question Answering. Methods Inf Med 56(3): 209-216.
- Afzal M, Hussain M, Malik KM, Lee S (2019) Impact of Automatic Query Generation and Quality Recognition Using Deep Learning to Curate Evidence from Biomedical Literature: Empirical Study. JMIR Med Inform 7(4): e13430.
- Prakash A, Saha SK (2019) A study on use of the web for automatic answering of remedy finding questions of common users. Technol Health Care 27(1): 23-35.
- Blair Goldensohn S, Mckeown K, Schlaikjer A (2003) A hybrid approach for QA track definitional questions. Proceedings of the 12th Text Retrieval Conference Maryland, pp. 185-192.
- Xu JX, Weischedel R, Weischedel A (2004) Evaluation of an Extraction-Based Approach to answering definitional questions. SIGIR’04, Sheffield, South Yorkshire, UK, pp. 418-424.
- Navigli R, Velardi P (2010) Learning word-class lattices for definition and hypernym extraction. Proceedings of the 48th Annual Meeting of the Association for Computational Linguistics, Sweden, pp. 1318-1327.
- Qian F, Yuan C (2012) A definition extraction algorithm combining hard pattern matching and soft pattern matching. Computer Technology and Development 22(9): 32-36.
- Storrer A, Wellinghoff S (2006) Automated detection and annotation of term definitions in German text corpora. Lrec, pp. 2373-2376.
- Gaudio R, Branco A (2007) Automatic extraction of definitions in Portuguese: a rule based approach. Proceedings of the 13th Portuguese Conference on Artificial Intelligence EPIA, pp. 659-670.
- Cui H, Kan M, Chua T (2004) Unsupervised learning of soft patterns for generating definitions from online news. Proceedings of WWW, New York, USA, p. 90-99.
- Cui H, Kan M, Chua T (2005) Generic soft pattern models for definitional question answering. Proceedings of the 28th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval (SIGIR ’05), ACM Press, New York, pp. 384-391.
- Cui H, Kan M, Chua T (2007) Soft pattern matching models for definitional question answering. ACM Transactions on Information Systems 25(2): 1-30.
- Borg C, Rosner M, Pace G (2009) Evolutionary algorithms for definition extraction”. Proceedings of the International Workshop on Definition Extraction, p. 26-32.
- Espinosa Anke L, Saggion H (2014) Applying dependency relations to definition extraction”. Springer International Publishing 8455: 63-74.
- (2021) SNOMED International. SNOMED CT Starter Guide.
- Bostock M, Ogievetsky V, Heer J (2011) D3: Data-Driven Document. IEEE transactions on visualization and computer graphics 17(12): 2301-2309.
- Heer J, Bostock M (2010) Declarative language design for interactive visualization. IEEE transactions on visualization and computer graphics 16(6): 1149-1156.
- Zhang YL, Zhang ZF, Zhang XD, Xu DS (2013) Web Dynamic Interactive Visualization of Knowledge Organization Systems with D3.js. New Technology of Library and Information Service 29(7): 127-131.

 Research Article
Research Article