ABSTRACT
We earlier identified four Gram positive Cr(VI) reducing bacteria (SUCR44, SUCR140, SUCR186 and SUCR188) isolated from tannery effluent irrigated soil and established that Cr(VI) reduction activity was localized in cell free extracts (CFE) rather than cell lysates (CL). In this paper we optimized the kinetics parameters (Km and Vmax) and the effect of pH and temperature on kinetics of CFE. The optimal temperature and pH for Cr(VI) reduction by CFE of aforesaid strains were 28°C and 7.0, except for SUCR188, which required lower temperature (20–28°C) and pH (5.0–6.0) optima. The maximum specific activity of Cr(VI) reduction was observed to be 0.42, 0.56, 0.45 and 0.49 μmol Cr(VI) min-1 mg-1 protein for strains SUCR44, SUCR140, SUCR186 and SUCR188, respectively. At their respective optimal temperature and pH, the minimum Km and maximum Vmax was found in the CFE of SUCR140.
Keywords: Cell Free Extracts; Cr(VI); Cr(VI) Reductase; Specific Activity
Introduction
Cr(VI), a widespread pollutant, is released into the environment by several industrial applications. It does not stay to the site of initial contaminant due to its soluble nature. Cr(VI) exists in solution as CrO4 2- and due to structural similarity with SO4 2-, can overcome the cellular permeability barrier, entering via sulphate transport pathways (Patra, et al. [1]), rapidly reducing to Cr(V) and generating free radicals (Mabbett, et al. [2]). The toxic properties of Cr(VI) originate from itself as an oxidizing agent as well as from the formation of free radicals. It is toxic (Wise, et al. [3]) to all forms of living systems causing oxidative stress (Ackerley, et al. [4]), DNA damage (Mabbett, et al. [2]) and altered gene expression (Bagchi, et al. [5]). Moreover, Cr(VI) is also mutagenic (Puzon, et al. [6]), carcinogenic (Codd, et al. [7]), and teratogenic (Asmatullah, et al. [8]), and has been recognized as a priority pollutant (Cheung, et al. [9]). Although hexavalent chromium is highly toxic, its trivalent form is relatively inert and much less toxic than the hexavalent form (Krishna, et al. [10]). Metal pollutants are non-degradable and can only be transformed to less toxic oxidation states or removed either by adsorption/accumulation or by physicochemical treatments. However, it has been observed that these processes are costly and unreliable (Malik [11]).
On the other hand, microbial reclamation is safe, ecofriendly and cost effective technology and an alternative to the traditional physicochemical methods. Several bacteria possessing chromate reductase activity have been reported, with ability to reduce Cr(VI) to Cr(III), which is much less toxic and less soluble, and thus reduction by these enzymes affords a means of chromate bioremediation. Our earlier studies conducted with four Gram positive chromate reducing bacteria (Soni, et al. [12]) indicated that the chromate reducing activity is associated with soluble fraction of cells which might be released extracellularly also. Despite the optimal conditions required for growth of bacteria, external pH and temperature (abiotic factors) condition may vary. However, the bacterial cells maintain their internal pH at around neutral. The pH homeostasis of the cells may be maintained by plasma membranes using the Na+/H+ antiporter system, K+/H+ antiporter, and ATPase–driven H+ expulsion (Horikoshi, et al. [13,14]). As the aforesaid abiotic factors can vary greatly in the environment, affecting the ability of microorganisms to reduce pollutants, knowledge of the kinetic factors is necessary for the designing an efficient bioremediation treatment for Cr(VI). This paper presents the results of our experiment conducted to study the effect of environmental factors like pH and temperature on kinetic parameters for Cr(VI) bioreduction by a crude cell free extracts of four Gram positive bacteria found efficient in reduction of chromate in our earlier studies.
Materials and Methods
Preparation of Cell Free Extracts
Cell-free extracts of bacterial isolates were prepared following previously published protocol (Soni, et al. [12]). Cells grown for 18 h in 250 ml Nutrient broth (5 g Sodium chloride l-1, 1.5 g Beef extract l-1, 1.5 g Yeast extract l-1, 5 g Peptic digest of animal tissue l-1, pH 7.0 ± 0.2) (Himedia, India) were harvested (OD at 600 nm were 1.2 ± 0.1) by centrifugation at 6,000 × g for 10 min at 4 °C, washed and resuspended in 20 ml of 0.1 M potassium phosphate buffer pH 7.0. These cell suspensions were placed in ice bath and disrupted using an Ultrasonic Probe (Rivotek, frequency 30 KHz ± 3 KHz) at 120 W with 15 second pulses at 15 second interval for 30 min. Sonicates thus obtained were then ultracentrifuged at 175,000 × g (Beckman coulter) for 90 min at 4 °C. The cytosolic fractions or supernatants thus obtained were filtered through 0.22 μm filters to yield the cell-free extracts devoid of membrane fractions and were immediately used for Cr(VI) reduction assay.
Enzyme Assays
Chromate reduction was estimated by using a standard calibration curve of Cr(VI) as in the form of K2CrO4. The reaction system (of 1 ml) used, contained varying Cr(VI) final concentrations (50–500 μmol) in 0.7 ml of 0.1 M potassium phosphate buffer (pH 7.0) with 0.3 ml aliquots of cell-free extracts for chromate reduction. The system volume of 1 ml was kept constant for all experiments. Assay conditions were kept constant with a reaction time of 30 min at 28 °C. Abiotic control contained corresponding concentration of Cr(VI) in 0.7 ml of phosphate buffer (0.1 M) with 0.3 ml of heat (100 °C for 30 min) treated cell free extract. Experiments for all isolates were done in triplicates. Unit enzyme activity for chromate reductase was derived as amount of enzyme that reduces 1 mM of Cr(VI) per min at 28 °C. Specific activity was defined as unit chromate reductase activity milligram-1 protein concentration in the cell-free extract. The residual Cr(VI) in cell free extract were estimated by 1,5–Diphenylcabazide method described by APHA (1995). Protein concentrations of cell-free extract were estimated using Folin-phenol reagent by reading absorbance at 750 nm, following the principle of (Lowry, et al. [15]). Known concentrations of Bovine serum albumin (BSA) prepared in phosphate buffer (pH 7.0) were used for drawing the standard calibration curve.
Effect of pH and Temperature on Cr(VI) Reduction by Cell-Free Extracts
Chromium reduction by CFE was studied at different pH (5.0, 6.0, 7.0, 8.0 and 9.0) and temperatures (20, 28, 35, and 42 °C) at 0.2 mM Cr(VI) concentration. The effect of pH on the reduction of Cr(VI) by the cell-free extracts of different SUCR strains was determined by using various buffers (50 mM sodium acetate, pH 4.0–5.5; 50 mM sodium phosphate, pH 5.5–8.0; 50 mM sodium carbonate, pH 8.0–10.0). The effect of temperature was determined by incubating the reaction mixtures for 30 min at different temperatures. Heat killed cell-free extracts treated at 100 °C for 30 min were used to check non-enzymatic reduction in respective strains. In our previous experiments (Soni et al. 2013) we observed that SUCR cells performed better at pH 7.0 and 28 °C temperature. So the temperature of 28 °C and a pH of 7.0 were taken as constants for studying the effect of different pH and temperature respectively.
Determination of Kinetic Parameters
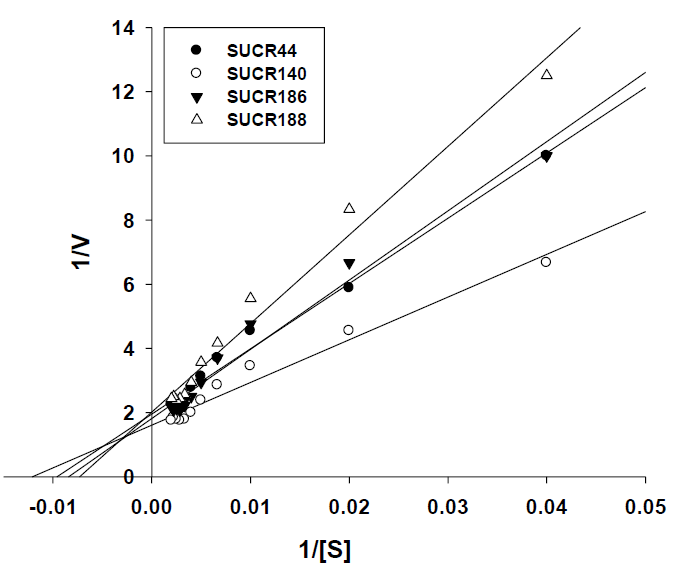
The enzyme kinetics was studied using the enzymatic progress curve using specific activity of chromate reduction by the cell free extracts. The kinetic constants were calculated by fitting the initial rate data to a double-reciprocal Lineweaver–Burk plot of 1/V [μmol Cr(VI) min-1 mg-1 protein] versus 1/[Cr (VI)] (μmol L-1) derived from a linear transformation of the Michaelis–Menten equation. This allowed the estimation of the specific Km and Vmax for cellfree extract reduction. Sigma Plot 10 software was employed for plotting the graphs.
Results and Discussion
Effect of Temperature and pH on Cr(VI) Reductase Activity
The Cr(VI) reducing strength of cell free extracts was found to be affected by strains identity, temperature and pH. (Tables 1a & 1b). Significant interactions were noticed for aforementioned parameters. The maximum chromate reductase activity in the cellfree extracts of all four strains at 0.2 mM Cr(VI) was established at 28 °C (Figure 1). Similar temperature optima (28–30 °C) for Cr(VI) reduction has been reported in other Gram positive bacteria including Bacillus (Camargo, et al. [16-20]). However, bacterial chromate reductase, active and stable at high temperature, has also been isolated from Thermus scotoductus found to be active and stable between 50–80 °C and was not active at low temperatures (Opperman, et al. [21]). At the temperature optima of 28 °C, the specific activity of Cr(VI) reduction was determined to be 0.32, 0.42, 0.34 and 0.28 μmol Cr(VI) min-1 mg-1 protein for strains SUCR44, SUCR140, SUCR186 and SUCR188, respectively. Considering Cr(VI) reductase activities as 100%, the Cr(VI) reductase activity of SUCR44, SUCR140, SUCR186 and SUCR188 decreased at lower temperature of 20 °C by 31% (0.22 μmol Cr(VI) min-1 mg-1 protein), 51% (0.205 μmol Cr(VI) min1 mg-1 protein), 32% (0.23 μmol Cr(VI) min-1 mg-1 protein) and 5% (0.265 μmol Cr(VI) min-1 mg-1 protein), respectively.
Figure 1: Effect of Temperature on chromate reduction activity by cell free extracts of different SUCR strains at pH 7.0 for 30 min incubation.
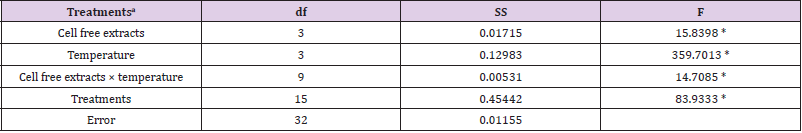
Table 1a: Summary of statistical analysis: the main effect and interaction of cell free extracts and temperature on Cr(VI) reduction were analyzed by factorial ANOVA.
Note: aCell free extracts of strains (SUCR44, SUCR140, SUCR186 and SUCR188); temperature (20 °C, 28 °C, 35 °C and 42 °C).
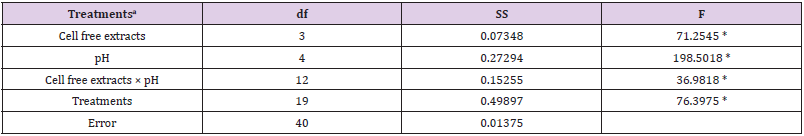
Table 1b: Summary of statistical analysis: the main effect and interaction of cell free extracts and pH on Cr(VI) reduction were analyzed by factorial ANOVA.
Note: aCell free extracts of strains (SUCR44, SUCR140, SUCR186 and SUCR188); pH (5.0, 6.0, 7.0, 8.0, 9.0).
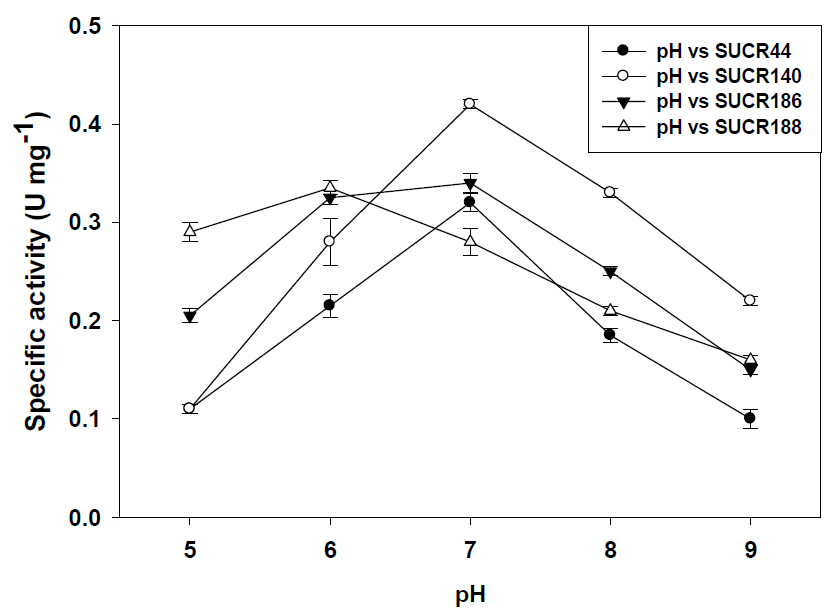
Similarly, assays with crude cell-free extracts at 35 °C showed a decrease of 41% (0.19 μmol Cr(VI) min-1 mg-1 protein), 61% (0.165 μmol Cr(VI) min-1 mg-1 protein) 31% (0.235 μmol Cr(VI) min-1 mg-1 protein) and 57% (0.12 μmol Cr(VI) min-1 mg-1 protein) of aforesaid strains respectively. At 42 °C the cell-free extracts of SUCR44, SUCR140, SUCR186 and SUCR188 retained 33% (0.105 μmol Cr(VI) min-1 mg-1 protein), 20% (0.085 μmol Cr(VI) min-1 mg-1 protein), 35% (0.12 μmol Cr(VI) min-1 mg-1 protein) and 20% (0.055 μmol Cr(VI) min-1 mg-1 protein) of the Cr(VI) reductase activity. These results indicate that amongst all four strains Cr(VI) reduction in SUCR44 and SUCR186 was least affected by changes in temperature. Assays with heat killed cell-free extracts (100 °C for 30 min) did not exhibit any chromate reductase activity in any of the said strains. In general, the activity of chromate reduction decreased at both alkaline and acidic pH. The optimum pH and temperature have been earlier observed to be the range (pH 5.0–9.0 and temperature 28–30 °C) reported for bacterial chromate reductases (Camargo, et al. [16,22]). The effect of pH on Cr(VI) reduction by cell free extract was determined at pH range of 5.0–9.0. The optimum pH for Cr(VI) reduction by the cell free extract at 0.2 mM Cr(VI) concentration, higher specific activities were found to be at pH 7.0 for SUCR44, SUCR140 and SUCR186, whereas, SUCR188 showed maximum Cr(VI) reductase activity at pH 6.0.
Specific Cr(VI) reductase activity at respective optimal pH for strains SUCR44, SUCR140, SUCR186 and SUCR188 was observed to be 0.32, 0.42, 0.34 and 0.37 μmol Cr(VI) min-1 mg-1 protein respectively (Figure 2). Considering these activities of respective strains as 100%, the relative effect of pH was determined. At pH 5.0., the specific activity of Cr(VI) reduction in SUCR44, SUCR140, SUCR186 and SUCR188 decreased by 66% (0.11 μmol Cr(VI) min-1 mg-1 protein), 74% (0.11 μmol Cr(VI) min-1 mg-1 protein), 41% (0.2 μmol Cr(VI) min-1 mg-1 protein) and 11% (0.33 μmol Cr(VI) min-1 mg-1 protein), similarly at pH 6.0 the 33% relative decrease were observed in both SUCR44(0.22 μmol Cr(VI) min-1 mg-1 protein), and SUCR140 (0.28 μmol Cr(VI) min-1 mg-1 protein), while no significant decrease was observed in SUCR186. At pH 7.0, the Cr(VI) reduction in SUCR188 decreased by 24% (0.28 μmol Cr(VI) min-1 mg-1 protein). At pH 8.0, 58% (0.185 μmol Cr(VI) min-1 mg-1 protein), 79% (0.33 μmol Cr(VI) min-1 mg-1 protein), 74% (0.25 μmol Cr(VI) min-1 mg-1 protein) and 57% (0.21 μmol Cr(VI) min-1 mg-1 protein) of specific activity was retained, respectively by aforesaid strains. Decrease in Cr(VI) reduction activity by 31% (0.1 μmol Cr(VI) min- 1 mg-1 protein), 48% (0.22 μmol Cr(VI) min-1 mg-1 protein), 56% (0.15 μmol Cr(VI) min-1 mg-1 protein) and 57% (0.16 μmol Cr(VI) min-1 mg-1 protein) were observed at pH 9.0 by strains SUCR44, SUCR140, SUCR186 and SUCR188 respectively.
Figure 2: Effect of pH on chromate reduction activity by cell free extracts of different SUCR strains at 28 oC and 30 min incubation.
Similar, observations of the influence of pH on bacterial Cr(VI) reduction have been made by others (Pal, et al. [17-19]). These results indicate that amongst all four strains Cr(VI) reduction in SUCR188 was least affected by changes in pH. Our results also suggest that cell free extracts of all the bacteria included in our study performed best at around neutral pH (except for SUCR188) required for the maximal growth of their cells too (Soni, et al. [12]). SUCR188 although is a mesophilic bacterium, being isolated from tannery effluent irrigated soil with an optimum growth of pH 7.0 (Soni, et al. [12]), the optimal pH for chromate reduction activity by crude cell free extract was 5.0–6.0. These results suggest the possibility of application of the crude enzyme in detoxification of Cr(VI) having moderate acidic pH condition, whereas resting cells of SUCR188 are more suitable for Cr(VI) sites with neutral pH.
Effect of Initial Concentration of Cr(VI) on Cell Free Extracts
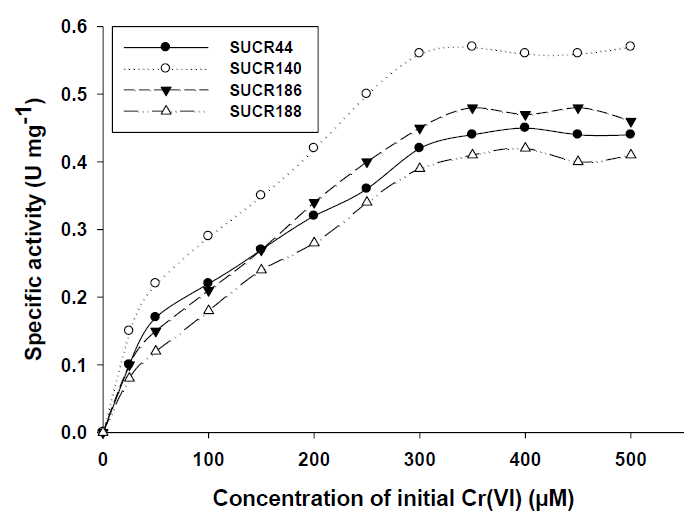
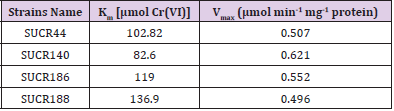
The effect of initial concentration of Cr(VI) on reductase activity of cell free extract was determined at a concentration range of 50– 500 μM of Cr(VI). An increase in the specific activity of chromate reduction by cell-free extracts of all the four bacteria was noticed with an increase in the initial concentration of Cr(VI) from 0 to 300 μmol, beyond which the activity was almost stationary (Figure 3). At optimal temperature and pH for respective strain, the observed maximum specific activity of Cr(VI) reduction for SUCR44, SUCR140, SUCR186 and SUCR188 at 300 μM were 0.42, 0.56, 0.45 and 0.49 μmol Cr(VI) min-1 mg-1 protein respectively (Figure 3). The kinetics of Cr(VI) reductase activity fitted well with the linearized Lineweaver- Burk plot (Figure 4), and thus the Km and Vmax values obtained. The calculated apparent Km and Vmax of different SUCR strains are shown in (Table 2). At a temperature and pH optima of respective strains the maximum Vmax and minimum Km was observed by SUCR140 followed by SUCR 44, SUCR186 and SUCR188. The Km and Vmax of SUCR44, SUCR186 and SUCR188 differed from the cell free extracts of other Bacillus sp. such as Bacillus firmus KUCr1 (Sau, et al. [23]), Bacillus sp. (Elangovan, et al. [17]), B. sphaericus AND303 (Pal, et al. [16]), Bacillus sp. ES29 (Camargo, et al. [15]), B. subtilis (Garbisu, et al. [24]). Lower Km values suggest higher affinity of the cell free extracts for the substrate [25]. Although a lot of work has been carried out on kinetics of cell free extracts of Bacillus sp., to the best of our knowledge this is the first report on kinetics study of Cr(VI) reduction by cell free extract of a Microbacterium sp.. [26]
Figure 3: Kinetics of Cr(VI) reduction by cell free extracts of SUCR strains. Reaction times was 30 minute.
Figure 4: Lineraized Lineweaver-Burk plot for Cr(VI) reduction of cell free extracts of different SUCR strains.
Conclusion
In conclusion, the cell free extracts of SUCR140 showed maximum Vmax and lowest Km values among all the four bacterial species included in the study. Although cell free extract of SUCR44, SUCR140 and SUCR186 performed best at pH 7.0 and 28 °C, also found optimal for their growth, SUCR188 performed better at moderate acidic and comparatively lower temperature. The generated information may be useful in selecting the strains vis -avis sites for improved remediation of chromium in eco-friendly way.
Acknowledgment
The authors wish to thank the Director, CSIR-Central Institute of Medicinal and Aromatic Plants, Lucknow, India, for providing necessary facilities and encouragement during the course of investigation and the Indian Council of Medical Research (ICMR), New Delhi, India, for providing financial support to SKS.
References
- Patra RC, Malik B, Beer M, Megharaj M, Naidu R, et al. (2010) Molecular characterization of Chromium (VI) reducing potential in Gram positive bacteria isolated from contaminated sites. Soil biol biochem 42(10): 1857-1863.
- Mabbett AN, Macaskie LE (2001) A novel isolate of Desulfovibrio sp. with enhanced ability to reduce Cr (VI). Biotechnol Lett 23(9): 683-687.
- Wise SS, Elmore LW, Holt SE, Little JE, Anto nucci PG, et al. (2004) Telomerase mediated lifespan extension of human bronchial cells does not affect hexavalent chromium induced cytotoxicity or genotoxicity. Mol Cell Biochem 255(1-2): 103-112.
- Ackerley DF, Barak Y, Lynch SV, Curtin J, Matin A, et al. (2006) Effect of chromate stress on Escherichia coli K-12. J Bacteriol 188(9): 3371-3381.
- Bagchi D, Stohs SJ, Downs BW, Bagchi M, Preuss HG, et al. (2002) Cytotoxicity and oxidative mechanism of different forms of chromium. Toxicology 180(1): 5-22.
- Puzon GJ, Petersen JN, Roberts AG, Kramer DM, Xun L, et al. (2002) A bacterial flavin reductase system reduces chromates (III) - NAD+ complex. Biochem Biophy Res 294(1): 76-81.
- Codd R, Irwin JA, Lay PA (2003) Sialoglycoprotein and carbohydrate complexes in chromium toxicity. Curr Opi Chem Biol 17(2): 213-219.
- Asmatullah Qureshi SN, Shakoori AR (1998) Hexavalent chromium induced congenital abnormalities in chick embryos. J Appl Toxicol 18(3): 167-171.
- Cheung KH, Gu JD (2007) Mechanism of hexavalent chromium detoxification by microorganisms and bioremediation application potential: A review. Int Biodeterior Biodegrad 59(1): 8-15.
- Krishna RK, Philip L (2005) Bioremediation of Cr(VI) in contaminated soils. J Hazard Mater 121(1-3): 109-117.
- Malik A (2004) Metal bioremediation by growing cells. Environ Int 30(2): 261-278.
- Soni SK, Singh R, Awasthi A, Singh M, Kalra A, et al. (2013) In vitro Cr(VI) reduction by cell-free extracts of chromate-reducing bacteria isolated from tannery effluent irrigated soil. Environ Sci Pollut Res 20(3): 1661-1674.
- Horikoshi K (1999) Alkaliphiles: Some applications of their products for biotechnology. Microbiol Molecul Biol Rev 63(4): 735-750.
- In: Horikoshi K, Antranikian G, Bull AT, Robb FT, Stetter KO, et al. (Eds.)., (2011) Extremophiles handbook, (1-2) Springer, Berlin.
- Lowry OH, Roseberough NJ, Lewis AF, Randall JR (1951) Protein measurement with the folin phenol reagent. The J Biol Chem 193(1): 265-275.
- Camargo FAO, Bento FM, Okeke BC, Frankenberger WT (2003) Chromate reduction by chromium-resistant bacteria isolated from soils contaminated with dichromate. J Environ Qual 32(4): 1228-1233.
- Pal A, Dutta S, Paul AK (2005) Reduction of hexavalent chromium by cell free extract of Bacillus sphaericus AND 303 isolated from serpentine soil. Curr Microbiol 51(15): 327-330.
- Elangovan R, Abhipsa S, Rohit B, Ligy P, Chandraraj K, et al. (2006) Reduction of Cr(VI) by a Bacillus sp.. Biotechnol Lett 28(4): 247- 252.
- Desai C, Jain K, Madamwar D (2008) Evaluation of In vitro Cr(VI) reduction potential in cytosolic extracts of three indigenous Bacillus sp. Isolated from Cr(VI) polluted industrial landfill. Biores Technol 99(14): 6059-6069.
- Ibrahim SS, El Tayeb MA, Elbadawi YB, Al Salamah AA, Garabed Antranikian, et al. (2012) Hexavalent chromate reduction by alkaliphilic Amphibacillus sp. KSUCr3 is mediated by copper-dependent membrane-associated Cr(VI) reductase. Extremophiles 16: 659-668.
- Opperman DJ, Piater LA, Heerden E (2008b) A Novel Chromate Reductase from Thermus scotoductus SA-01 Related to Old Yellow Enzyme. J Bacteriol 190(8): 3076-3082.
- Bae WC, Lee HK, Choe YC, Jahng DK, Lee SH, et al. ( 2005) Purification and characterization of NADPH dependent Cr(VI) reductase from Escherichia coli ATCC 33456. J Microbiol 43(1): 21-27.
- Sau GB, Chatterjee S, Mukherjee SK (2010) Chromate reduction by cell-free extract of Bacillus firmus KUCr1. Polish J Mrobiol 59(3): 185-190.
- Garbisu C, Alkorta I, Llama MJ, Serra JL (1998) Aerobic chromate reduction by Bacillus subtilis. Biodegradation 9(2): 133-141.
- APHA (American Public Health Association); American Water Works Association (AWWA); Water Environment Federation (WEF) (1995) Standard Methods for the Examination of Water and Wastewater (19th)., Washington, DC
- Park CH, Keyhan M, Wielinga B, Fendorf S, Matin A, et al. (2000) Purification to homogeneity and characterization of a novel Pseudomonas putida chromate reductase. Appl Environ Microbiol 66(5): 1788-1795.

 Research Article
Research Article