Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Stéphanie Aguero1*, Simon Megy1, Aurélie Thibaut2, Frédéric Delolme2, Virginie Gueguen Chaignon2 and Raphaël Terreux1
Received: January 24, 2023; Published: February 08, 2023
*Corresponding author: Stéphanie Aguero, Équipe ECMO, Laboratoire de Biologie Tissulaire et d’Ingénierie (LBTI), UMR5305,Université Lyon 1, Lyon, France
DOI: 10.26717/BJSTR.2023.48.007658
Cytochromes P450 are multifunctional enzymes which usually act as mono-oxygenases. However, the CYP450 Thaxtomin E has been shown to carry out a direct nitration reaction on the aromatic residue L-Tryptophan. The details of the related mechanism by which TxtE generates ONOO° radicals through its ferric complex have been partially elucidated. We investigated the possible role of Tyr89 in the radical nitration of L-Trp into 4-L-nitro-Trp. Tyr89 is involved in a network of hydrogen bonds with residues of the FG loop and is directly positioned above the C4 of the L-Trp bound substrate. On this basis, we hypothesized that Tyr89 could act as an intermediate for the nitroso radical before the final nitration. We first performed quantum calculations to evaluate the possibility of a direct or indirect nitration. Then we produced a Y89F mutant to disactivate the potential role of the tyrosine on the radical nitration. Binding and nitration tests finally validated one of our hypotheses. Our results show that the nitro radical transfer by Tyr89 is possible in terms of enthalpy and geometry. However, the experimental nitration assays of the Y89F mutant allows us to conclude that this is a direct radical reaction and that Tyr89 is not involved in the process.
Keywords: Nitration; Radical Reaction; Thaxtomine E; 4-L-nitrotryptophan; Quantum Calculations; Computational Chemistry; Binding Assay Experiment; Enzymatic Activity Assay; Mass Spectrometry
Abbreviations:Txt: Thaxtomin; 4-L-nitro-Tryptophan: 4-L-nitroTrp; CYP450: Cytochrome P450; L-Tryptophan: L-Trp; NADPH: Nicotinamide Adenine Dinucleotide Phosphate; CcO: Cytochrome C Oxidase; DFT; Density Functional Theory; MSM: Markov State Model: RNS: Reactive Nitrogen Species; ROS: Reactive Oxygen Species; DEANO: Diethylamine NONOate
In biochemical terms, the aromatic nitration process introduces a nitro group (-NO2) into aromatic cycles. This mechanism is found in various biological systems and is usually a physiological operation. However, under some circumstances, this process can sometimes lead to pathological consequences. In vegetal pathology, this reaction is involved in the common scab, a disease that causes significant lesions on the tuber surface of the plant. In potatoes, the pathology is induced by the bacteria Streptomyces Scabies, which produces a phytotoxin referred as Thaxtomine A (TxTA), which is responsible for the plant cell hypertrophy and seedling stunting [1-3]. The TxtA toxine contains a nitro group that is essential for its phytotoxicity [4]. TxTA synthesis is a multi-step operation and is carried out by six Txt enzymes: TxtA, TxtB, TxtC, TxtD, TxtE, and TxtR [4]. It has been shown that TxtA phytotoxine is formed by the assembly of L-phenylalanine and L-4-nitrotryptophan (4-L-nitroTrp) by a nonribosomal peptide synthetase [5]. Even though the biosynthesis of the nonproteinogenic amino acid 4-L-nitroTrp is not yet fully understood, it has been established that TxtE, the thaxtomin responsible for this reaction, is a cytochrome P450 (CYP450).
CYP450 are heme-dependent enzymes which catalyze many reactions related to biodegradation and biosynthesis [6]. They usually act as mono-oxygenases, as they transfer one atom of molecular oxygen to a substrate, while the second oxygen atom of the O2 molecule is reduced into water [7]. However, some families can perform other various reactions, including nitration [8]. TxtE belongs to a novel P450 subfamily, which is able to regiospecifically nitrate aromatic amino acids. Indeed, TxtE produces 4-L-nitro-Trp from L-tryptophan (L-Trp) using nitric oxide, oxygen, and Nicotinamide adenine dinucleotide phosphate (NADPH) [9]. The details of the catalytic mechanism have remained unclear for a long time. A potential mechanism, based on the general mechanism of CYP450 and the substrates used and associated with the products of the reactions, has been proposed by (Barry, et al. [4]). L-Trp would bind to the active site of TxtE, triggering the loss of the water ligand from the ferric heme. This first step is in adequation with the observed spin state change. The binding would promote NADPH-dependent reduction of the heme to its ferrous form by ferredoxin and ferredoxin reductase. Then, O2 would bind the heme iron ion, resulting in a ferric superoxide complex. Finally, NO would react with the ferric superoxide complex, resulting in a ferric peroxynitrite complex. Peroxynitrite could undergo a homolytic cleavage into the NO2 and an iron (IV)=O species (often referred to as ‘compound II’). Recently, (Louka, et al. [10]) have shown that the reaction actually proceeds through an iron (II)-nitrosyl intermediate, in which dioxygen binds first to the heme iron followed by NO to generate the iron (III)-peroxynitrite species (Figure 1).
Regarding the nitration of the bound L-Trp, several hypotheses have been proposed. Nitration could occur via NO2 addition and compound II-mediated hydrogen atom abstraction, resulting in an iron (III)- OH species. An alternative mechanism for nitration would involve a protonation-triggered heterolytic cleavage of the ferric peroxynitrite complex to yield NO2+ and an iron (III)-OH species, followed by a classical electrophilic aromatic substitution [4]. However, Louka, et al. [10] recently showed that the reaction of NO and O2 on an iron (III)-heme generates free NO2• radicals. On this basis, we explored the nitration phase independently of the NO2 generation mechanism. We used the hypothesis of a NO2• radical generator as a starting point. The nitration of L-Trp is commonly described as direct, as no experimental evidence of reaction intermediates are available. In the case of a radical reaction, as no reaction intermediates are observable, the assumption of a direct reaction cannot be validated a priori. However radical reactions are known to have a fast-kinetic mechanism, suggesting that the reaction is most likely a direct type of reaction. Louka et al. confirmed that point with quantum calculations. Here we explore a different approach, based on experimental observations: the free radical nitration mediated by Tyr89. We propose a second mechanism that would involve the formation of a nitro-tyrosine intermediate. Our hypothesis is based on the mechanism of the CcO cytochrome c oxidase, which is the terminal enzyme in the respiratory chain reducing molecular oxygen to water [11]. The active site of this enzyme has, apart from its bi-nuclear heme-copper center, a highly conserved tyrosine residue, which is also found in the active binding site of TxtE. Its role is to deliver both an electron and a proton during the cleavage step of the O - O bond, forming a tyrosyl radical. Then, the catalytic cycle takes place in four stages of reduction. One study suggests that in three of them, one proton of the tyrosine interacts with the bi-nuclear center, leaving the unprotonated tyrosine in a radical state. Tyrosine protonation would occur in the final reduction step. This tyrosine-linked reduction mechanism is believed to be essential for the functioning of the enzyme. Density functional theory (DFT) calculations confirmed this theory, which is consistent with independent experimental observations [12].
We analyzed the active site of TxtE, which also contains a tyrosine residue, as observed in the cytochrome c oxidase enzyme. In a substrate bound conformation [13], L-Trp is positioned in the active site in a tight orientation, due to hydrogen bond interactions with several amino acids, including Arg59, Tyr89, Thr296, Asn293 and Glu394. The double cycle is secured by hydrophobic interaction with Met 88 and Phe395. A sequence alignment highlighted the highly conserved amino acids in the TxtE enzyme, confirming that Tyr89 is a highly conserved residue. (Dodani, et al. [13,14]) showed by molecular and experimental simulation methods that this tyrosine plays a fundamental role in the binding of the substrate. Starting from the L-Trp-bound form (PDB ID 4TPO), they generated about 100 molecular dynamics which they analyzed using an approach based on the Markov State Model Theory (MSM) [15]. The latter discretizes the conformational set in a collection of states and builds a matrix with the transition probabilities between them. This analysis evidenced that the FG loop acts as a lid and coexists in 2 different states: open and closed. In the closed lid state (active conformation). the substrate indole ring of L-Trp can occupy the active-site pocket in two distinct orientations, as illustrated in Figure 2:
In the ‘unflipped’ state (A), the indole NH is in contact with the heme-bound peroxynitrite oxygen, in accordance with what is observed in the co-crystal structure, and Tyr89 is located close to the C4 of the L-Trp. In the ‘flipped’ state (B), the indole ring is rotated by ∼180° with its C4 closest to the nitrogen atom of the heme-bound peroxynitrite and Tyr89 is located in opposition to the nitration site. We hypothesize that in the ‘unflipped’ orientation (A), Tyr89 could contribute to the nitration of the C4 by acting as a reactional intermediate receiving the NO2• radical. Indeed, the formation of 3-nitrotyrosine is a process observed in the context of oxidative stress, involving Reactive Nitrogen Species (RNS) derived from nitric oxide (NO•) and the superoxide ion (O2•-). Tyrosine is not exclusively nitrated in position 3, as many intermediate states exist [16].
Figure 3. Mechanism of 4-nitrotyrosine formation.
a) Tyrosine residue at the start of the reaction
b) Oxydized Tyrosine by ROS compound
c) Tautomeric equilibrium with (b)
d) Expected reaction, but not observed because of the steric hindrance
e) Intermediate nitro compound
f) The direct attack by the NO2
• is not energetically favorable.
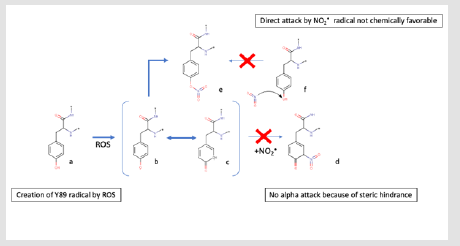
Figure 4. Rearrangement of Arg59 following the anchoring of L-Trp in the active site. The L-Trp is displayed in cyan, and the residues Trp82, Tyr89 and Arg59 respectively in magenta, green and yellow. The residue Arg59 from the 4TPN crystal (TxtE alone) has been added in blue to underline its conformational change consecutive to the L-Trp binding.
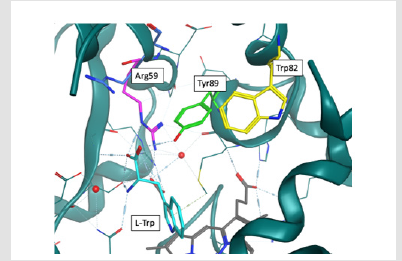
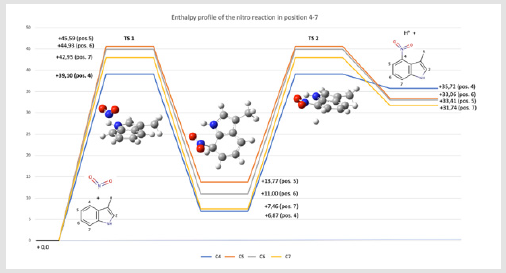
Figure 5.Energy of the reaction of the nitration. All the calculations were computed with UM062X hybrid method with 6-311++G(d,p) basic set. All the computed energies are expressed in kcal.mol-1.
On the basis of these elements, we thus hypothesized a potential role of Tyr89 intermediates in a possible mechanism of radical nitration. In this scenario, the NO2• radical would no longer directly attack L-Trp but Tyr89, whose OH function would be previously radicalized by a reactive oxygen species (ROS) compound (Figure 3), thus forming a 5-O-nitrotyrosine. The formation of such a transient state has never been observed with the TxtE protein but has been experimentally confirmed [17]. 4-Nitrotyrosine, instead of being converted into 3-nitrotyrosine as observed by (Radi, et al. [18]), would attack L-Trp in position 4. This hypothesis is based on the comparison between the 4TPO (TxtE linked to L-Trp) and 4TPN (TxtE only) structures. Upon binding of L-Trp, the formation of a hydrogen bond between the amino acid Arg59 and the carboxylate group of L-Trp causes the αB’1 helix reorganization, which leads to the anchoring of the substrate in the catalytic position [14]. This rearrangement displaces Arg59 relatively close to Tyr89. Sterically, the formation of a 3-nitrotyrosine would pose a hindrance problem. Likewise, a 5-nitrotyrosine isomer would not be possible because of the presence of Trp82 in opposition to the C5 of Tyr89 (Figures 4 & 5). The nitration reaction of L-Trp by the TxtE enzyme constitutes the key step in the biosynthesis of TxtA. Although the NO-bound nitration process is not uncommon [19], TxtE is the first known enzyme to catalyze a specific direct nitration reaction in a biosynthetic pathway. Its potential for direct application in the industrial sector is therefore promising. The goal of this paper is to verify our hypothesis by a combined experimental and computational study. First quantum calculations were performed in order to evaluate the possibility of a direct or indirect nitration. Then a Y89F mutant was experimentally produced to disactivate the potential tyrosine effect on radical nitration. Binding and nitration tests finally validated our hypothesis.
Quantum Calculations
All the computational studies were carried out using the density functional theory (DFT) methods implemented in the Gaussian 09 D.01 suite of programs [20]. The equilibrium geometries of the molecules were determined using the gradient technique. The force constants and vibrational frequencies were determined by computing analytical frequencies on the stationary points obtained after optimization in order to check if there were true minima. The basis set used in this work was standard 6-311G++(2d, p). For the calculation of the molecular structure and properties of the studied system, we chose the hybrid meta-GGA density functionals M06- 2X [21], which consistently provides satisfactory results for several structural and thermodynamic properties [21-23].
Cloning, Protein Expression and Purification
The full-length TxtE DNA sequence from S. scabies 87.22 (406 amino acids) (TxtE-wt) and the TxtE-Y89F mutant were cloned into pET-28a plasmids with a N-terminal 6His tag. Cloning was performed by the Twist Bioscience company. The purification work benefited from the SFR Biosciences Protein Science Facility (http://www. sfr-biosciences.fr). E. coli BL21star-DE3 (Novagen) bacteria were transformed, grown in LB media (Sigma-Aldrich) to OD600=0.6 and the expression was induced with 0,5 mM IPTG at 18 °C overnight. Cells were harvested after a 15 min of centrifugation at 5,000xg and resuspended in a lysis buffer containing 25 mM Tris pH 8, 300 mM NaCl, 10% glycerol, 0,1 mg/ml lysozyme, 5 μg/mL DNase, 5 mM MgCl2 and 1 mM DTT. Disruption of the cells was achieved with sonication after addition of a home-made antiproteases mix. Cell debris were removed by centrifugation for 20 min at 20,000xg at 4 °C. The recombinant protein was purified by chromatography using the AKTA Pure system (Cytiva) and a Histrap HP prepacked column (Cytiva). Unbound material was extensively washed using 25 mM Tris pH 8, 300 mM NaCl, 20 mM Imidazole, 1 mM DTT and 10% glycerol. Then both TxtE proteins (TxtE-wt and TxtE-Y89F) were eluted using a 25 to 300 mM imidazole gradient over 8 column volumes. Peak fractions were pooled, and proteins were further purified using an anion exchange column (HiTrap Q HP - Cytiva). Finally, the purification of the TxtE proteins was completed using a size exclusion column (Superdex 75 increase 10/300, Cytiva) equilibrated with 25 mM Tris pH 8, 150 mM NaCl, 5% glycerol. The purity of the sample was assessed by SDS-PAGE. Freshly purified TxtE proteins were concentrated at 5 mg/ml with 10 KDa Amicon Ultra concentrators (Millipore). Purified proteins were aliquoted, flash frozen and stored at -80 °C until further use.
L-Trp Binding Assays of TxtE
Purified TxtE-wt and TxtE-Y89F proteins were desalted and diluted to a concentration of 15 μM in the final buffer (25 mM Tris pH 8), using a Zeba spin desalting column (Thermofisher Scientific). Proteins were plated in 96-well plates (Greiner bio-one) in the presence or not of 1 mM L-Trp (Sigma–Aldrich). Absorbance spectra were recorded between 325 and 500 nm using a UV-vis spectrophotometer (Infinite TECAN M1000) according to (Yu, et al. [9]).
TxtE-Catalyzed Nitration Reaction
Both proteins were desalted on a Zeba spin column and diluted to 50 μM in 25 mM Tris pH 8. The other components of the nitration reaction were prepared as follow: Diethylamine NONO ate sodium salt hydrate (DEANO, Sigma–Aldrich) was prepared at 100 mM in 10 mM sodium hydroxide and further diluted to 100 μM in 25 mM Tris pH 8. NADPH (Sigma–Aldrich) was freshly prepared at 18 mM in 25 mM Tris pH 8. Ferredoxine (Fd, Sigma–Aldrich) was resuspended to 1 mg/ml in 25 mM Tris pH 8. Ferredoxine NADP+ reductase (FR, Sigma–Aldrich) was purchased at 1 U/mL in 35 mM Tris, 0,1 M NaCl, 20% glycerol. L-Trp was prepared in water at 10 mM. The nitration reaction was performed as described by (Dodani, et al. [13]), in a final volume of 200 μL in 25mM Tris pH 8 containing 1,5 μM TxtEwt or TxtE-Y89F, 0,5 mM L-Trp, 5 μM DEANO, 1 mM NADPH, 2 μg Fd and 0,034 U FR. The reaction was incubated for 2 hours at room temperature. Each reaction was filtered using a 0,5 mL 3-kDa MWCO unit (Millipore). The flow-through was transferred to a vial insert and frozen at -20 °C until further analysis by liquid chromatography mass spectrometry (LC-MS).
Mass-Spectrometry
Samples were analyzed on an Ultimate 3000 nano-RSLC (Thermofisher Scientifics, San Jose California) coupled online with a Q Exactive HF mass spectrometer via an IonFlex nano-electrospray ionization source (Thermofisher Scientifics, San Jose California) set at 1.9 kV. First, samples were diluted in water (1/50) and injected on a C18 Acclaim PepMap100 trap-column 75 μm ID x 2 cm, 3 μm, 100 Å, (Thermofisher Scientifics) via the full loop (1 μl) injection mode. Then, an isocratic elution was performed for 5.0 minutes at 2 μL/min with 2% acetonitrile, 0.05% trifluoroacetic acid in water. Full scan MS data were acquired at a resolution of 240,000 at m/z 200 Th. The mass range was set to m/z 130-400, the Ion Target Value to 3E6 and the maximum injection time was set to 100 ms. For the data analysis, the eXtracted Ion Chromatograms (XIC) of m/z 205.0977 (tryptophan) and m/z 250.0828 (Nitrotryptophan) were plotted and areas ATryptophan and ANitrotryptophan were obtained after integration of the signals. The ratio ATryptophan / ANitrotryptophan was then calculated.
Computational Calculations
Direct Nitration by Free-Radical Addition. Quantum Study of the Direct Nitration Mechanism of L-Trp to 4-L-Nitro-Trp:In this hypothesis, the nitro radical (NO2•) would attack the carbons 4 to 7. The targeted aromatic carbon, which possesses a sp2 hybrid configuration, would be modified into a tetrahedral sp3 configuration, resulting in the partial loss of aromaticity of the cycle. In a second step, a radical would be created in the alpha position of the addition. The system would then recover its aromaticity with the release of a H• radical. We computed the variation of enthalpy of the reaction involving the nitro radical attack on positions 4 to 7 using the hybrid method. The tetrahedral intermediate was optimized for each position, using a scan and transition search algorithm for the transition state conformation. All the calculations were performed using the UM06-2X method with the atomic base 6-311++ G (d, p) using the Gaussian 09D software in gas phase. For the transition state structure, the frequency was computed to verify the real intermediate state with the negative frequency vibration. The sum of the electronic and thermal enthalpies was extracted from each structure and the ΔHr was computed. The transition state 1 (TS1), which corresponds to the attack of the nitro radical (NO2•) compound on the C4 has the lowest enthalpy energy (+ 39,1 kcal.mol-1) starting from the tryptophan residue and the nitro radical. The intermediate compound corresponds to the reactive carbon with a sp3 configuration. Then a H• radical is released (TS2) and returns to the heme to reduce iron IV to III. The calculations agree with the literature and the experimental observations [4].
Free Radical Nitration Mediated by Tyr89. Quantum Study of the Direct Nitration Mechanism of L-Trp to 4-L-Nitro-Trp: In the case of the nitration by Tyr89, the hydroxy function would react with a free radical species to form a hydroxy radical (TyrO•). The nitro radical would be released from the central heme and would be bonded with the hydroxy radical. Then the nitro radical would be transferred to the substrate according to the reaction described above. This assumption was investigated with the same protocol as previously described. The formation of the hydroxy radical has a ΔHr of 0.1370 ha or 85.99 kcal.mol-1, which is absorbed by the reaction by the ROS. It should be noted that the attack of the nitro radical (NO2•) in alpha of the hydroxy function is impossible due to steric hindrance with the residues Arg59 and Trp82. Then the nitro radical would react with the hydroxyl radical to form a bond. The ΔHr of the reaction is -0.0383 ha or -24.08 kcal.mol-1. The formation of a bond between TyrO• and NO2• is direct without any transition state. Then, once the tryptophan is stabilized in the site, the NO2• radical could be released with a cost of +24.08 kcal.mol-1. We thus conclude that, based on the quantum calculation, in terms of enthalpy and geometry, the nitro radical transfer by Tyr89 is entirely acceptable and possible.
Experimental Data
L-Trp Binding Assays of TxtE: Substantial amounts of TxtE-WT and TxtE-Y89F mutant proteins were obtained when overexpressed in E. coli BL21star-DE3. They were purified to homogeneity as described in the Materials and Methods section. After a desalting step, their ability to bind tryptophan was first tested and validated with a spectrophotometric assay. Their absorbance spectrum were recorded and a maximum of absorbance at 420 nm was observed for both TxtE-WT and TxtE-Y89F, as previously described [9]. Because TxtE is a P450 cytochrome, the binding of tryptophan is observed with a blue shift of the maximum of absorbance, both for TxtE-WT and TxtEY89F. This results in a characteristic type-I binding spectrum arising from the conversion of the heme iron atom from low to high spin upon tryptophan binding [4]. Therefore, the Y89F mutation does not affect the tryptophan binding on the TxtE protein. Subsequently, the ability of TxtE-WT and TxtE-Y89F enzymes to metabolize L-Trp into 4-nitro-L-Trp was assessed. The nitration reaction was carried out with the purified enzymes mixed with L-Trp substrate, a NO donor, NADPH, ferredoxin and ferredoxin reductase, for 2 hours at room temperature. Control samples were prepared with boiled TxtE (95 °C for 5 minutes), or without one of the reagents (L-Trp or Diethylamine NONOate (DEANO) or ferredoxin). L-Trp in solution was also analyzed in presence or in absence of DEANO. The samples were then analyzed by liquid chromatography coupled with mass spectrometry [13].
Table 1.L-Trp nitration assay of TxtE-wt and TxtE-Y89F. ATryptophan and ANitrotryptophan represente areas of the eXtracted Ion Chromatograms (XIC) of m/z 205.0977 (Tryptophan) and m/z 250.0828 (Nitrotryptophan). The relative percentage of ATryptophan and ANitrotryptophan is also calculated. Different conditions are tested for the TxtE-WT and the Txt-Y89F: (1 and 6) The mix is composed of the purified enzymes TxtE-wt or TxtE-Y89F with L-Trp substrate, DEANO (NO° donor), NADPH, Ferredoxin and Ferredoxin reductase; (2 and 7) mix without L-Trp; (3 and 8) mix without Ferredoxin reductase; (4 and 9) mix without DEANO; For the controls: (5 and 10) boiled mix, which represents the negative control; (11) mix without protein; (12) L-Trp only; (13) L-Trp with DEANO only.
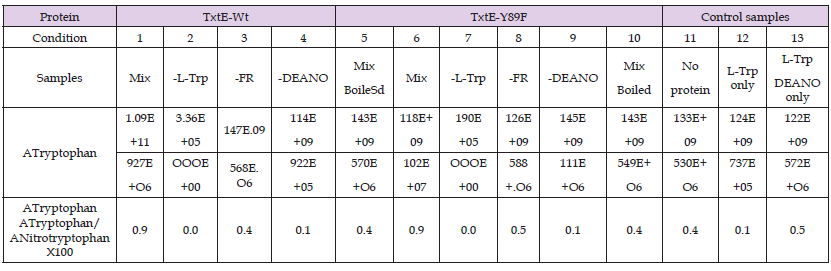
MS Results: All the Calculated ratios ANitrotryptophan / ATryptophane are shown in Table 1. We tested different conditions for each protein. The ratio was first measured for the complete system (Table 1, conditions 1 and 6) composed of the NO° donor DEANO, NADPH, ferredoxin, ferredoxin reductase, and L-Trp. Then, we alternately removed the substrate, ferredoxin, or DEANO. Calculated ratios ANitrotryptophan / ATryptophane show the absence of nitration when the NO donor (DEANO) is absent (Table 1, conditions 4 and 9). Indeed, the nitration ratio drops to 0.1. This value corresponds to the basal nitration of L-Trp due to contamination (Table 1, condition 12). For the complete systems (Table 1, conditions 1 and 6), we observe a nitration ratio of 0.9. When the ferredoxin reductase is omitted (Table 1, conditions 3 and 8), the ratios drop respectively to 0.4 for Txt-WT and 0.5 for the TxtE-Y89F mutant. When the Txt-WT or TxtE-Y89F protein is denatured (Table 1, conditions 5 and 10), the nitration ratio also drops to 0.4. This value of 0.4 is entirely resulting from the chemical nitration of DEANO (see control Table 1, condition 13: L-Trp w. DEANO). We conclude that the nitration level remains the same in the samples containing no or only L-Trp. When DEANO is present, the nitration level is increased by an approximate factor of 10. In the absence of the purified proteins or FR, the nitration still occurs, despite the nitration ratio being divided by 2. No difference is observed between TxtE-wt and TxtE-Y89F. The nitration is entirely due to the enzyme. The mechanism of the TxtE protein is radical, and therefore is not dependent on Y89.
Louka, et al. [10] demonstrated that the formation of peroxynitrite by the TxtE protein leads to a NO2• radical generation. They also used quantum calculations to decribe the entire L-Trp nitration mechanism as part of a direct attack of NO2• on the C4 of L-Trp. Our study explores this direct radical attack from another angle. It has been shown that peroxynitrite promotes the nitration of tyrosine residues in some proteins through a process catalyzed by transition metals. For example, (Ischiropoulos, et al. [24]) discovered that peroxynitrite leads to nitration within Manganese Superoxide Dismutase. The overall mechanism of the tyrosine nitration in vivo is therefore also based on the chemistry of free radicals. Free-radical chemistry provides high-energy intermediates. Tyrosine does not react directly with peroxynitrite, but instead with radicals derived from the latter and other single-electron oxidants, leading to the formation of the tyrosyl radical (Tyr•), which, in turn, reacts with NO2• to give the final stable product, 3-nitrotyrosine [17]. The study of crystals of TxtEWT co-crystallized with L-Trp [13] shows that Tyr89 is positioned above the C4 of the L-Trp. By relying on this data set, we studied the potential role of Tyr89 in the TxtE mechanism. First, we performed quantum calculations to explore the radical nitration of the tyrosine as an intermediate of the reaction. Then we implemented experimental methods in order to verify our hypothesis: first, we used DFT to study the different intermediates of the reaction. The calculated energies suggest that direct nitration is as likely as indirect nitration by Tyr89. These results are not surprising given the nature of the reaction. Radical reactions are known to cause chain reactions, and tyrosine is an amino acid able to undergo radicalization.
Using MSM simulations, (Dodani, et al. [14]) showed that the substrate indole ring of L-Trp can be found within the active-site pocket in two distinct orientations: ‘unflipped’ and ‘flipped’ as illustrated in Figure 2, the indole ring being rotated by ∼180°. We hypothesized that in the ‘unflipped’ orientation (Figure 2A), Tyr89 could contribute to the nitration of the C4 by acting as a reactional intermediate receiving the NO2• radical. For a closed-lid state, the ‘unflipped’ orientation exposes the C4 in close proximity to Tyr89. This conformation could allow the indirect nitration pathway. Their calculations evidenced that the unflipped orientation is observed in 22% of the time, which would be sufficient for the indirect nitration process to occur. In order to validate this hypothesis, we decided to express a mutant Y89F protein, where the ability of Tyr89 to produce a radical function through its OH terminal group would be disactivated, thanks to the mutation into phenylalanine. Our quantum study results demonstrate that this hypothesis is clearly possible in terms of energy. However, our binding and nitration assays show that the mutation of Tyr89 into Phe89 does not prevent the L-Trp nitration. As a result, we conclude that Tyr89 is not involved in the nitration process, as the nitration rates are similar with the wild-type enzyme and the Y89F mutant.
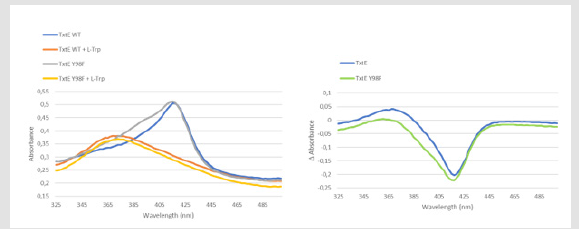
On a separate note, the precise role of Tyr89 can also be discussed. It should be noted that (Dodani, et al. [14]) proposed that Tyr89 would be involved in the binding of L-Trp through a network of H-bonds, as observed in their crystal (PDB accession code 4TPO). Our experimental results with the mutant protein show that the binding is not affected, as we observe similar profiles for the L-Trp binding (Figure 6). This raises questions on the exact role of Tyr89. However, upon the Y89F mutation, the two binding curves (TxteE Y89F + L-Trp and TxtE W + L-Trp) show similar profiles with a slight downward shift of the absorbance. This slight shift can very well be explained considering that Tyr89 is not essential for the stabilization of the substrate, but contributes in a synergically way to the binding and the stabilization of the L-Trp. Finally, we ask the question of a possible role in the regioselectivity of the reaction. Its position just above the C4 and the fact that Tyr89 is a highly conserved residue are substantial clues about a potential role during the reaction. As we demonstrated that Tyr89 is not involved in the nitration process itself, the next question is naturally its role in the regioselectivity. The MS analysis we provided did not aim to identify the different enantiomers but quantify the presence of nitro L Trp versus L-Trp. A more in-depth study would allow us to highlight and quantify the presence of possible enantiomers and, therefore, further explore the role of Tyr89 in this mechanism
Figure 6.L-Trp binding assay for TxtE-wt and TxtE-Y89F: a. UV-visible spectra of TxtE-wt and mutant with or without L-Trp; b. Differential UVvisible spectra.
The enzymatic mechanism of the TxtE protein has been well-studied for several years. Its potential for agricultural and pharmacological applications is extremely promising, and the engineering of the TxtE protein could lead to numerous potential applications in the industry as well. Although its mechanism has been partially elucidated, some gray areas still remain. We decided here to explore the nitration mechanism from a new angle by assuming that, in some cases, the nitration would involve Tyr89 as a nitration intermediate. When the FG loop is closed, the L-Trp substrate is mostly found in a ‘flipped’ state, positioning the C4 of L-Trp in the direction of the heme center donor of the NO2• radical. We hypothesized a possible nitration mechanism taking advantage of the ‘unflipped’ state under the same conditions. In this case, Tyr89, an amino acid known for its ability to undergo radical nitration, which is positioned in opposition to the C4, would contribute to the nitration of L-Trp by transiently supporting the NO2• radical before its release to L-Trp. Our quantum analysis shows that this mechanism is quite credible and to a certain extent as realistic as the direct mechanism. The production of the TxtE Y89F mutant protein allowed us to test and verify whether this hypothesis was tangible. However, our experimental results lean more towards the hypothesis of a direct nitration mechanism. This study, at last, allowed us to study and nuance the possible role of Tyr89 in the binding of the substrate within the active site.
Conceptualization, S.A., S.M. and R.T.; methodology, S.A., S.M., V.G.C., F.D., A.T. and R.T.; software, S.A, S.M., F.D. and R.T.; validation, S.A., S.M., V.G.C., F.D. and A.T.; formal analysis, S.A., S.M., V.G.C., F.D. and A.T.; resources, S.A. and S.M.; data curation, S.A. and S.M.; writing— original draft preparation, S.A., S.M., V.G.C., F.D. and A.T.; writing— review and editing, S.A., S.M. and R.T.; visualization, S.A, S.M., V.G.C. and F.D.; supervision, S.M. and R.T.; project administration, S.M., V.G.C., F.D. and R.T. All authors have read and agreed to the published version of the manuscript.
This research received no external funding.
The authors declare no conflict of interest.


