Impact Factor : 0.548
- NLM ID: 101723284
- OCoLC: 999826537
- LCCN: 2017202541
Lalita Kaushal, Kumari Alka, Arti and Duni Chand*
Received: July 20, 2023; Published: August 14, 2023
*Corresponding author: Duni Chand, Department of Biotechnology, Himachal Pradesh University, Shimla, Himachal Pradesh, India
DOI: 10.26717/BJSTR.2023.52.008215
Introduction: Liver disorders that poses serious threat to human life and account for approximately two
million worldwide deaths per annum. Extensive research is still going on to develop and discover novel and
safer drug molecules with lesser side effects on human health. In-silico methods not only decrease the cost
and time of the drug discovery but also make developmental process faster and reliable.
Objective: The present research aimed to identify hepatoprotective and other pharmaceutical properties
of traditionally used medicinal plant Thalictrum foliolosum.
Material and Methods: In this study 15 phytochemicals present in the plant were used for the molecular
modelling and in-silico activity prediction. Six protein targets involving in liver disorder were considered
for the docking study and performed using PyRx Software.
Results: Among 15 phytochemicals thalicarpine and dibenzylisoquinoline were shown to inhibit the
proteins TNF-α and TGF-β and their binding affinity toward TNF-α and TGF-β were more than silymarin i.e.,
-11.8 Kcal/mol, -9.1 Kcal/mol, -8.6 Kcal/mol, -13.2 Kcal/mol, -10.3 Kcal/mol and -9.8 Kcal/mol respectively.
Thalicarpine was shown to result in activation of the proteins PPAR-α, PPAR-γ and PPAR-β/δ with binding
affinity more than silymarin i.e., -9.5 Kcal/mol, -12.4 Kcal/mol and -11.9 Kcal/mol respectively. Thalicarpine
and 1, 3 Dibenzylisoquinoline showed more affinity and interaction toward Autophagy inducers protein
than silymarin i.e., -15.7 Kcal/mol and -11.3 Kcal/mol respectively.
Conclusion: In-silico activity prediction studies revealed the great potential of these phytochemicals like
antioxidant, anti-protozoal, anti-leukemic, and anti-viral and cystic fibrosis treatment etc.
Keywords: Hepatoprotective; Polypharmacology; Bioinformatics, Molecular Docking
Abbreviations: TNF-α: Tumor Necrosis Factor- α; TGF-β: Transforming Growth Factor- β; PPAR- α: Peroxisome Proliferator-Activated Receptor- α; PPAR- γ: Peroxisome Proliferator-Activated Receptor- γ; PPAR-β/δ: Peroxisome proliferator-activated receptor- β/δ
Being the second largest organ in the human body, liver not only plays censorious role in the synthesis of bile, blood clotting factor, metabolism of nutrients, drugs, xenobiotics but also act as a detoxifying organ by modulating biotransformation for the removal of toxic and waste substances from the body (Huang, [1,2]). Liver lobule, the basic anatomical and functional unit of the liver is composed of hepatic cords with central and portal veins at each end where two types of epithelial cells are present i.e., hepatocytes; a type of parenchylmal cells involved in the functional properties of the liver and cholangiocytes which are bile duct lining cells (Huang, Qiang He, et al. [1-3]). Liver is the only organ in human body that is well known for remarkable regenerative capacities of the hepatocytes which is vital for the maintenance of liver function during homeostasis and after liver damage. Regeneration power of the liver can be diminished by viral infection, drug toxicity, mycotoxin toxicity, chemical toxicity, alcohol consumption, chemotherapeutics side effects and carcinogenesis, resulting in irreversible damage to the liver (Huang, et al. [1-3]). Liver damage may pose a serious threat to human life so to reduce risk factors extensive research is ever going on to develop safer and less side effect causing drug molecules (Krishnan et al., 2017). Complementary and alternative medicine involves use of plant-based materials for treatment of various complications (Jinadatta, et al. [4]). Nowadays, the use of plants and plants-based products as food supplements, food additives and pharmaceutical agents has been rising around the globe. World Health Organization (WHO) has reported that almost 30 % of the plants and their extract somehow have been used as healthcare compounds (Fan Yi et al., 2018). Plant based medicines are economic, less toxic and exhibit more efficacy in case of multidrug resistance which is due to the presence of broad range of phytochemicals in plants (Kabir et al., 2016).
In India, a number of plant extracts have been traditionally used and claimed to treat liver disorders but scientific validation, safety conformity and mechanism of action is still lacking. Natural phytochemicals reduce the risk of hepatic damage caused by oxidative stress (Ellappan, et al. [2]). Thalictrum foliolosum was traditionally used for the treatment of jaundice, edema, atonic dyspepsia, and skin diseases (Salaria, et al. 2022). The ethano-pharmacological uses of the medicinal plants have well established history but scientific study and their mechanism of action determination holds great importance in the development of more effective and safe health care system (Fan Yi et al., 2018). Determination of the pharmacological basis of the traditionally used medicinal plants involves two types of approaches, a traditional method, and computational methods. Traditional methods of the discovery and development of drugs are costly and time-consuming process (Fan Yi et al., 2018; Ghosh et al., 2022) and hence to overcome this cumbersome process a poly pharmacology approach has been emerged. Poly pharmacology involves the interactions of a given drug or pharmacological compound with multiple targets, which can target single disease pathway or multiple diseases pathways in a consortia manner (Sucularli [5]). Computational methods involve use of Bioinformatics tools for the development of drugs such as discovery of target molecules and the interaction between target and drug molecules. Computational methods reduce the cost of the drug discovery and development process. Therefore, these methods are valuable for the prediction of the probable activities and targets of the various phytochemicals present in different plants. The present research is mainly focused on the determination of the interaction between TNF-α (tumor necrosis factor- α), TGF-β (Transforming growth factor- β), PPAR- α (peroxisome proliferator-activated receptor- α) PPAR- γ (peroxisome proliferator-activated receptor- γ), PPAR-β/δ (peroxisome proliferator-activated receptor- β/δ) and autophagy inducer protein with various phytochemicals present in Thalictrum foliolosum to identify potential inhibitors and liver regeneration and function promoting properties.
Bioinformatics/ computational tools
The computational facilities used to perform in-silico investigations of various phytochemicals present in Thalictrum foliolosum included PubChem(www.pubchem.com), Open Babel GUI (36), RCSB PDB PyRx software and discovery studio (Salaria, et al. 2022).
Ligand preparation
The chemical structures of the various phytochemicals present in Thalictrum foliolosum viz., berberine, thalurgosamine, thalrugosidine, thalirugidine, 8-oxyberberine, thalirugine, jatrorrihizine, thalicarpine, , palmatine, thalidasine, noroxyhydrastinine thalisopine (talysopine), 1,3 dibenzylisoquinoline, 1,benzylisoquinoline, o-methylthalibrine and silymarin (Standard) obtained from the PubChem database (www.pubchem.com) in conical SMILES format for in-silico prediction studies and in .sdf format for docking studies. The Open Babel GUI was used to convert the .sdf files of the phytochemicals and standards into PDB format and are further prepared using the Open Babel program.
Protein preparation
Proteins targets selected for the presented research study were TNF-α, TGF-β, PPAR- α, PPAR- γ, PPAR-β/δ and autophagy inducer protein. The 3-D structure of the target proteins were obtained from the protein data bank (PDB ID; 3d24, 1VJY, 5HYK, 3C58, 5Y7X and 5x8i respectively Figure 1). These six proteins were selected for the molecular docking studies with various phytochemicals present in Thalictrum foliolosum to identify potential inhibitors and liver regeneration and function promoting properties.
Molecular Docking
The molecular docking studies of the phyto ligands with the six protein targets were performed using PyRx Software. For molecular docking vina Perl script was used and one of the best conformations with the lowest docking energy was chosen. Discovery Studio was used to analyze the PDB complexes of the target proteins and phyto ligands, to study the interactions between these complexes.
In-silico study for the pharmacokinetics and bioactive phytochemical toxicity prediction
In-silico screening study of the 15 selected phytochemicals present in Thalictrum foliolosum and silymarin (standard) was done using web based online servers such as PASS online web resource (Institute of Biomedical chemistry (IBMC) Russia), ProTox (Charite University of Medicine, Institute of Physiology, Structural Bioinformatics Group, Berlin, Germany), admetSAR (Laboratory of Molecular Modelling and Design, Shanghai, China) and SwissADME (Molecular Modeling Group of Swiss Institute of Bioinformatics, Lausanne, Switzerland) (Kabir et al., 2016; Salaria et al. 2022).
Molecular Docking of Phytochemicals Presents in Thalictrum Foliolosum with Target Proteins
The molecular docking studies were used to assess the energy binding interactions between the molecules of bio-target proteins and phytochemicals selected for the study. 3-D structure of the phytochemicals and target proteins were downloaded from PubChem and RCBS-PDB database. The results of the molecular docking study were compared with standard drug silymarin, and the results are summarized in Table 1. Molecular docking results showed the standard drug silymarin has docking score for interaction with TNF-α (3d24), TGF-β (1VJY), PPAR- α (5HYK), PPAR- γ (3C58), PPAR-β/δ (547X) and autophagy inducer protein (5x8i) as -8.6 Kj/mol, -9.8 Kj/ mol, -7.1 Kj/ mol, -9.2 Kj/mol, -9.5 Kj/mol and -11.1 Kj/mol respectively. Among all the 15 phytochemicals, thalicarpine and 1,3 dibenzylisoquinoline showed docking score for interaction with TNF-α (3d24), TGF-β (1VJY), PPAR- α (5HYK), PPAR- γ (3C58), PPAR-β/δ (547X) and autophagy inducer protein (5x8i) as -11.8 Kj/mol, -13.2 Kj/ mol, -9.5 Kj/ mol, -12.4 Kj/mol , -11.9 Kj/mol, -15.7 Kj/mol and -9.1 Kj/mol, -10.3 Kj/ mol, -8.6 Kj/mol, -8.6 Kj/mol , -9.3 Kj/mol and -11.2 Kj/mol respectively results are summarized in Table 2. The molecular docking results showed that thalicarpine and 1,3 dibenzylisoquinoline have binding affinity greater than silymarin (standard drug). The results of docking studies of TNF-α are presented in the Figure 2, which is showing 3-D and 2-D crystal structures of TNF-α with ligands viz., Silymarin (A), Thalicarpine (B) and 1,3 Dibenzylisoquinoline (C) having binding affinity -8.6 Kcal/mol, -11.8 Kcal/mol and -9.1 Kcal/mol respectively. Silymarin was able to form 6 hydrogen bonds with TNF-α at amino acids GLN A: 390, LYS A: 394, SER C: 339, GLY C: 393, MET C: 421 and ASP C: 423. Other interactions were with amino acids VAL C: 340, HIS C: 341 and MET C: 421. TNF-α forms 2 hydrogen bonds with thalicarpine at amino acid SER C: 263 and GLY C: 393. 1,3 Dibenzylisoquinoline does not form hydrogen bonds with TNF-α, but active pocket of protein showed other interaction at amino acids GLU C: 205, PRO C: 206, LYS C: 208 and ARG C: 276 (Table 2).
Table 1: Molecular docking interaction of all 15 phytochemicals present in Thalictrum foliolosum and standard drug silymarin with target proteins.
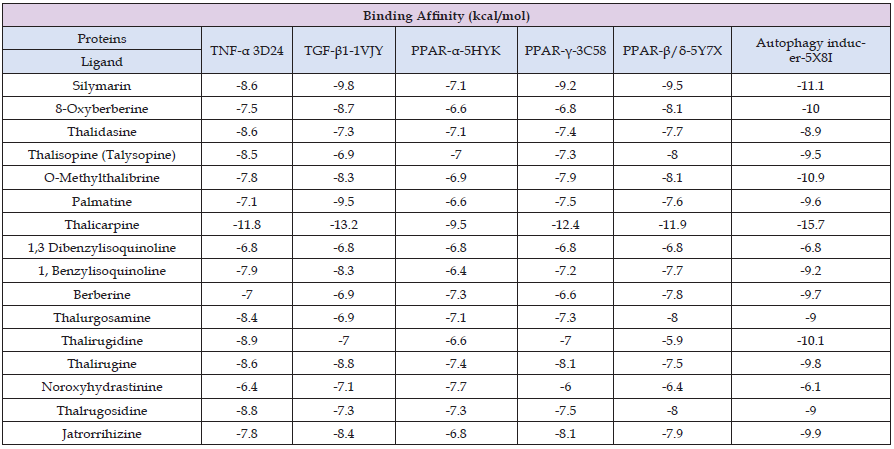
Table 2: Molecular docking analysis results of two best phytochemicals and standard drug silymarin with all six target proteins.

The results of the docking studies of TGF-β are presented in the Figure 3, which is showing 3-D and 2-D crystal structures of TGF-β with ligands viz., Silymarin (A), Thalicarpine (B) and 1,3 Dibenzylisoquinoline (C) having binding affinity -9.8 Kcal/mol, -13.2 Kcal/ mol and -10.3 Kcal/mol respectively. Hydrogen bonding’s and other molecular interactions in relation to TGF-β of thalicarpine were more than silymarin. Silymarin was able to form 2 hydrogen bonds with TGF-β at amino acids ARG A: 294 and HIS A: 285. Other interactions were with amino acids ASP A: 290, ILE A: 211, TYR A: 295, VAL A: 219, LEU A: 340 and ALA A: 230. TGF-β forms 3 hydrogen bonds with thalicarpine at amino acid ASP A: 290 and ASP A: 351. In the active pocket of the protein other interactions are found at amino acids LYS A: 337 and ASP A: 290. 1,3 Dibenzylisoquinoline does not form hydrogen bonds with TGF-β but active pocket of protein showed other interaction at amino acids ILE A: 211; VAL A: 219; ALA A: 230; LYS A: 232; LEU A: 260; TYR A: 282 and LEU A: 340 (Table 2). The results of the docking studies of PPAR- α are presented in the Figure 4, which is showing 3-D and 2-D crystal structures of PPAR-α with ligands viz., Silymarin (A), Thalicarpine (B) and 1,3 Dibenzylisoquinoline (C) having binding affinity -7.1 Kcal/mol, -9.5 Kcal/mol and – 8.6 Kcal/ mol respectively. Silymarin was able to form 2 hydrogen bonds with PPAR- α at amino acids ASN A: 326 and LEU A: 229. Other interactions were with amino acids VAL A: 240 and PHE A: 338. PPAR- α forms 1 hydrogen bond with thalicarpine at amino acid ASP A: 387. In the active pocket of the protein other interactions are found at amino acids ASP A: 387 and GLU A: 398. 1,3 Dibenzylisoquinoline form 1 hydrogen bond with PPAR- α at amino acid ASN A: 219, in active pocket of protein showed other interaction at amino acids THR A: 279; MET A: 320; VAL A: 324 and VAL A: 332 (Table 2).
The docking studies of PPAR-γ with ligands are presented in the Figure 5, which is showing 3-D and 2-D crystal structures of PPAR-γ with ligands viz., Silymarin (A), Thalicarpine (B) and 1,3 Dibenzylisoquinoline (C) having binding affinity -9.2 Kcal/mol, -12.4 Kcal/mol and -8.6 Kcal/mol respectively. Silymarin was able to form 3 hydrogen bonds with PPAR-γ at amino acids LYS A: 261, ARG A: 288 and SER A: 342. Other interactions were with amino acids CYS A: 285, ARG A: 288, ALA A: 292, LEU A: 330, LEU A: 333, ILE A: 341 and MET A: 348. PPAR- γ forms 2 hydrogen bonds with thalicarpine at amino acid LYS A: 261and ILE A: 281. In the active pocket of the protein other interactions are found at amino acids LYS A: 261, GLU A: 272 and PHE A: 287. 1,3 Dibenzylisoquinoline does not form hydrogen bonds with PPAR- γ but active pocket of protein showed other interaction at amino acids GLU A:259, GLY A:284, CYS A:285, ARG A:288 and ILE A:341 (Table 2). The results of the docking studies of PPAR-β/δ with ligands are presented in the Figure 6, which is showing 3-D and 2-D crystal structures of PPAR-β/δ with ligands viz., Silymarin (A), Thalicarpine (B) and 1,3 Dibenzylisoquinoline (C) having binding affinity -9.5 Kcal/ mol, - 11.9 Kcal/mol and -9.3 Kcal/mol respectively. Silymarin was able to form 6 hydrogen bonds with PPAR- α/γ at amino acids MET A: 192, TRP A: 228, ARG A: 248 and ASN A: 307. Other interactions were with amino acids PHE A: 190, CYS A: 251, MET A: 293 and ILE A: 297. PPAR- α/γ form 2 hydrogen bonds with thalicarpine at amino acid GLU B: 223. In the active pocket of the protein other interactions are found at amino acids GLU B: 223, LEU B: 235 and HIS B: 244. 1, 3 Dibenzylisoquinoline does not form hydrogen bonds with PPAR- α/γ but active pocket of protein showed other interaction at amino acids THR B: 252, ILE B: 290, MET B: 293, LEU B: 294 and ILE B: 297(Table 2).
The docking studies of Autophagy inducer protein are presented in the Figure 7, which is showing 3-D and 2-D crystal structure of Autophagy inducer protein with ligand Silymarin (A), Thalicarpine (B) and 1,3 Dibenzylisoquinoline (C) having binding affinity -11.1 Kcal/mol, -15.7 Kcal/mol and -11.3 kcal/mol respectively. Hydrogen bonding’s and other molecular interactions in relation to Autophagy inducer protein of thalicarpine were more than silymarin. Silymarin was able to form 2 hydrogen bonds with Autophagy inducer protein at amino acids LYS A: 290. Other interactions were with amino acids LEU A: 167; PHE A: 172; VAL A: 175; ALA A: 186; VAL A: 225; PHE A: 241; LEU A: 244; LEU A: 295 and VAL A: 324. Autophagy inducer protein forms 3 hydrogen bonds with thalicarpine at amino acid GLU B: 206, LEU B: 244 and ASN B: 293. In the active pocket of the protein other interactions are found at amino acids PHE B: 172, ASP B: 288 and ASP B: 325. 1,3 Dibenzylisoquinoline does not forms hydrogen bonds with Autophagy inducer protein, but active pocket of protein showed other interaction at amino acids PHE B: 172; VAL B: 175; ALA B: 189; VAL B: 225; VAL B: 324; ASP B: 325 and PHE B: 241 (Table 2). From the present study it is predicted that phytochemicals of Thalictrum foliolosum have pharmaceutical applications and potential to treat hepatic disorders.
In-Silico Study for the Pharmacokinetics and Bioactive Phytochemical Toxicity Prediction
In-silico PASS Prediction: The 15 phytochemicals present in Thalictrum foliolosum viz., berberine, thalurgosaminine, thalrugosidine, 8-oxyberberine, jatrorrihizine, noroxyhydrastinine, palmatine, thalicarpine, thalidasine, thalirugidine, thalirugine, thalisopine (talysopine), 1,3 dibenzylisoquinoline, 1, benzylisoquinoline, o-methylthalibrine were analyzed by the PASS online server for evaluation of their biological activities. Out of fifteen three phytochemicals namely palmatine, berberine and jatrorrihizine were not processed by the PASS online server rest of the compounds showed greater Pa than Pi (Table 3). 1, Benzylisoquinoline showed highest Pa values for antibacterial (0.291), antiviral (0.465), anti-protozoal (0.446) and cytochrome P450 stimulant (0.421) activities. 8-Oxyberberine showed the highest Pa values for antineoplastic (0.581), lipid peroxidase inhibitor (0.408) and TP53 expression enhancer (0.456) activities. Thalurgosaminine showed the highest Pa values for anti-leukemic (0.327) and TNF-α release inhibitor (0.354) activities. Thalrugosidine showed highest Pa values for antioxidant (0.218) and anti-inflammatory (0.455) activities. Thalidasine showed the highest Pa values for leukopoiesis stimulant (0.846) and cystic fibrosis treatment (0.392) activities. Thalisopine (talysopine), noroxyhydrastinine, 1, 3 dibenzylisoquinoline and o-methylthalibrine showed highest Pa values for free radical scavenger (0.258), hepato-protectant (0.230), anti-carcinogenic (0.218) and antifungal (0.103) activities. Pa (predicted activity) and Pi (predicted inactivity) values for all different activities of the examined phytochemicals are presented in Table 3.
In-Silico Studies Involving use of SWISS ADME, admetSAR and Protox-II Servers: Computational facilities of SWISS ADME, admet- SAR, and Protox-II servers were used to analyze pharmacokinetics and toxicities of phytochemicals having pharmacological relevance. Fifteen bioactive phytochemicals and a standard drug (silymarin) were selected for these studies. Using SWISSADME, all of the fifteen phytochemicals follow Lipinski’s rule of five. Whereas 13 phytochemicals follow the Veber’s rule and, phytochemicals thalirugidine, O-methylthalibrine and standard drug silymarin doesn’t follow Verber’s rule. Noroxyhydrastinine water soluble, 8-Oxyberberine, Berberine, Jatrarrhizine and standard drug were moderately soluble in water while rest phytochemicals were insoluble in water. The GI-absorption of all phytochemicals was predicted to be high, while silymarin predicted to have low GI-absorption. Using admetSAR, all the phytochemicals and silymarin predicted to be non-carcinogenic in nature. All the phytochemicals and silymarin were reported to be positive for human intestinal absorption except jatrarrhizine and berberine. Silymarin is negative for blood brain barrier penetration while all phytochemicals were positive for blood brain penetration and Caco-2 permeability. Protox- II server predicted LD 50 value for all fifteen phytochemicals ranges from 200 to 5000 mg/kg and for silymarin LD50 value was 2000 mg/kg. Organ toxicity study predicted that all the phytochemicals does not possess hepatotoxicity. Out of fifteen phytochemicals, five phytochemicals viz., thalurgosaminine, thalidasine, 8-oxyberberine, palmatine and berberine predicted carcinogenicity. All phytochemicals except noroxyhydrastinine, 1,3 dibenzylisoquinoline and 1, benzylisoquinoline predicted immnotoxicity. All phytochemicals except noroxyhydrastinine are predicted to have mutagenicity. Out of fifteen phytochemicals, three phytochemicals viz., 8-oxyberberine, palmatine and berberine predicted to have cytotoxicity. All the results are summarized in Table 4.
Table 4: Pharmacokinetics and toxicity prediction of the phytochemicals present in Thalictrum foliolosum.
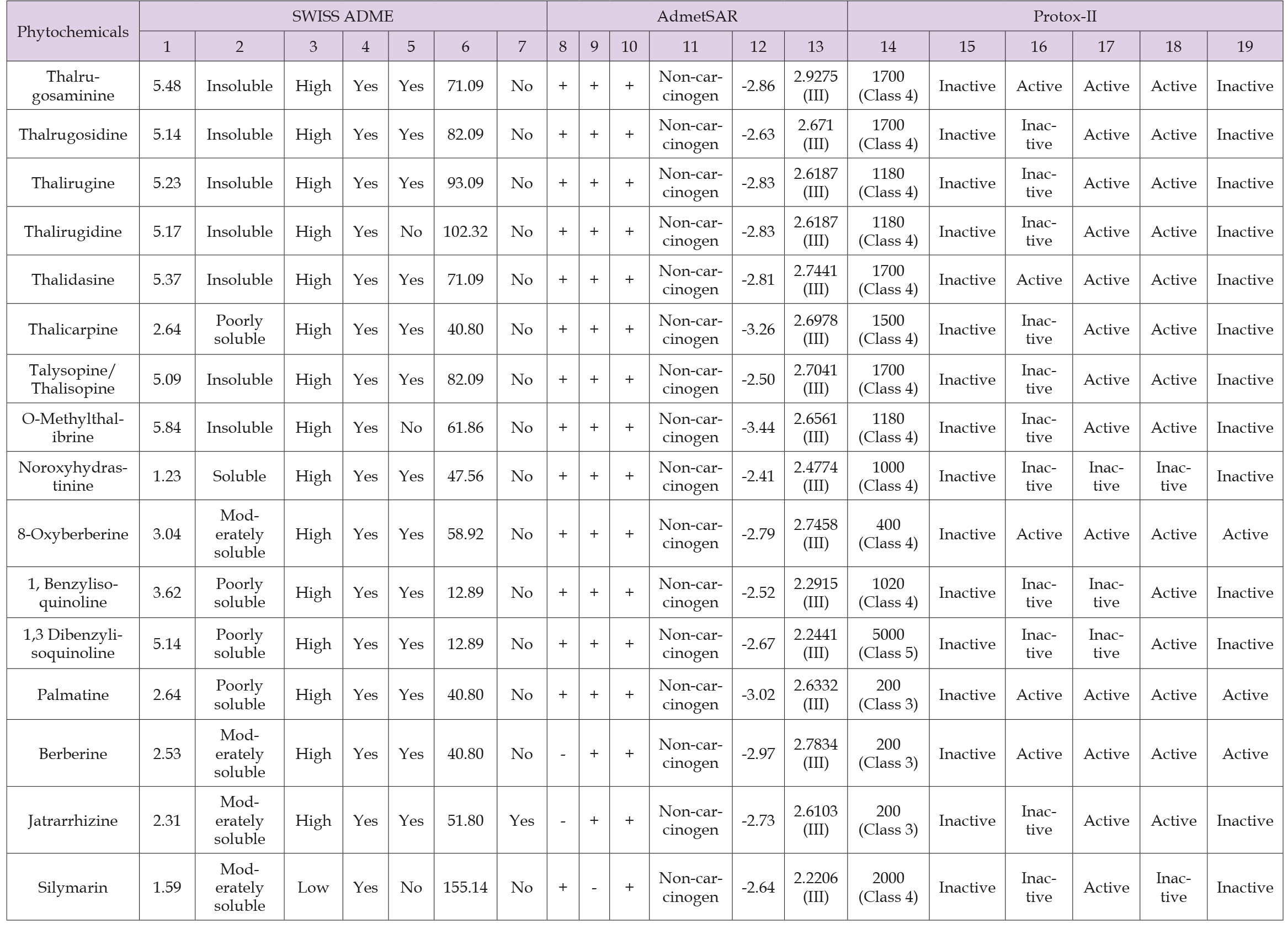
Note: Were
Consensus log P o/w.
Water Solubility
GI-absorption
Lipinski rule
Veber’s rule
TPSA (Å2)
Leadlikeness,
Human intestinal absorption
Blood brain barrier penetration
Caco-2 permeability
Carcinogen
Aqueous solubility (Log S value)
Rat acute toxicity (LD-50)
LD50 (mg/kg)
Organ toxicity hepatotoxicity
Carcinogenicity
Immunotoxicity
Mutagenicity
Cytotoxicity, positive ‘+’ negative ‘-’
Plant based herbal formulations have been used since time immemorial for the betterment of human health and are a desirable alternative to conventional drugs due to their high efficacy, cost effectiveness and easy availability (Daniyal et al., 2019). To explore the pharmaceutical potential of herbal formations and discovering novel pharmaceuticals extensive research is still going on to discover new drugs towards deadly diseases (Jinadatta, et al. [4]). Computational studies play a valuable role in the prediction of novel activities and targets for known compounds (Sucularli [5]). Liver disorders accounts for more than 2 million deaths annually and is the 14th number among the leading causes of deaths around the globe. Liver fibrosis causes excessive accumulation of the extracellular matrix components which leads to the production of TNF-α and TNF-α, pleiotropic cytokines, and are produced by different immune cells viz., macrophages/ monocytes. TNF-α is known to trigger multiple signaling pathways involved in proliferation, inflammation, and apoptosis. Liver fibrosis is further promoted by the pro-survival effect and pleiotropic effect of TNF-α on hepatic stellate cells (HSC). The initial event in the liver inflammation and fibrosis involves death of hepatocytes which is induced by TNF-α. Therefore TNF-α plays a salient role in the pro-inflammatory responses and in the cell-to-cell communication. TNF-α enhances hepatic stellate cells (HSC) survival, immune cells activation and hepatocyte death, all of these are associated with liver fibrosis. So, targeting TNF-α signaling pathway is regarded as a noble approach for the treatment of liver fibrosis (Yang and Seki, et al. [6-8]). Thalicarpine and 1, 3 Dibenzylisoquinoline were able to inhibit the protein TNF-α and their binding affinity toward TNF-α was more than silymarin. According to Ullah, et al. [9] poncirin treatment is known to attenuate expression of signaling proteins and inflammatory cytokines.
TGF-β plays a major role in chronic liver disease and is known to regulate all the stages of liver disorders as TGF-β is regulating intracellular pathways that induce liver injury and liver fibrosis (Jinadatta, et al. [4]). Thalicarpine and 1, 3 Dibenzylisoquinoline were able to impede the action of protein TGF-β and showed greater interaction towards TGF-β than silymarin (standard drug). Thus targeting TGF-β in peculiar cells at the specific time can help to achieve therapeutic effect on liver disorders. According to Jinadatta, et al. [4] gnetol is known to inhibit TGF-β and reported hepatoprotective effect of gnetol. PPAR-α participates in liver homeostasis and in various metabolic functions like regulation of lipid metabolism (Ellappan et al. Jinadatta et al. [2- 4]) and abnormalities related to PPAR-α may lead to liver cancer and hepatic steatosis. PPAR- α influences the severity of non-alcoholic liver disease and has anti-inflammatory properties as it counteracts nuclear factor kappa-B and enhances FGF21, the antitumor activity by regulation of nuclear factor kappa-B signaling. PPAR- α ligand exhibit anti-inflammatory, anti-steatotic and anti-fibrotic effect (Wang et al. [10]). PPAR-α can be activated by a few ligands. Thalicarpine and 1, 3 Dibenzylisoquinoline showed high affinity for binding and more efficacies through the interaction with PPAR- α protein than silymarin. According to Jinadatta, et al. 2019, gnetol results in the activation of PPAR- α protein which is a hepatoprotective ligand.
PPAR-γ plays a salient role in adipogenesis, metabolism of lipids, immune regulation, insulin sensitivities and other cellular processes. In liver cells it is known to regulate insulin receptors, glucokinase and sex hormone binding proteins. PPAR-γ is known to have anti-inflammatory properties regulating immune inflammatory response. Activation of PPAR-γ is known to suppress TNF-α so it could reduce hepatic fibrosis (Wang et al. et al. [1-10]). For PPAR-γ only thalicarpine was able to activate the protein PPAR-γ and its binding affinity towards PPAR-γ was more than silymarin. PPAR-β/δ activation may prevent dyslipidemia, obesity, insulin resistance, non-alcoholic liver disease and also increases liver glucose catabolism. PPAR-β/δ own anti-inflammatory effects in the liver as it inhibits NF-κB activity. For PPAR-γ only thalicarpine was able to activate the protein PPAR-γ and its binding affinity towards PPAR-γ was more than silymarin. Autophagy is an intracellular degenerative pathway targeting cytosolic components such as organelles and proteins into lysosomes to degrade these components for maintaining cellular functions and survival. Liver depends on autophagy for normal functioning as well as to prevent the development of diseases. Compounds inducing autophagy represent noble promising agents for the treatment of a diverse range of liver- related medical illnesses. However, safe autophagy-inducing compounds for clinical applications are still lacking (Czaja et al. [11]; Sun, et al. 2017). Thalicarpine and 1, 3 Dibenzylisoquinoline showed more affinity toward Autophagy inducers protein and interaction than silymarin. [12-26] According to Sun et al., 2017 compounds which are able to induce autophagy showed notable hepatoprotective effects in the mouse model against acetaminophen (APAP)-induced liver injury. Thus, targeting the above-mentioned protein targets in peculiar cells at the specific time will help to achieve therapeutic effect on most of the liver disorders.
The present studies provide in-silico evidence that support the pharmaceutical potential of bio-active phytochemicals present in Thalictrum foliolosum and support the use of these phytochemicals for the treatment of hepatic disorders, provide protection against bacteria, virus, fungus, protozoa, and free radical induced damage. Further studies for the validation of in-silico results and detailed in-vitro and in-vivo studies on pharmaceutical potential are also performed to support this study.
The author would like to express their sincerest gratitude to Professor Duni Chand for mentoring and supervision throughout the research work.
Conflict of Interest
No conflict of interest.
Author Contributions
LK was responsible for conceptualization, validation, formal analysis, investigation, writing original draft and visualization. KA and A were responsible for helping in the reviewing and editing. Professor DC was responsible for conceptualization, editing, supervision and reviewing the manuscript. All the authors contributed towards the elaboration of the manuscript and have given their approval of the final version.
Funding
The author is grateful to ICMR, New Delhi for providing funding to Lalita Kaushal in the form of JRF/SRF from 2017 to 2021.


