Abstract
The CRISPR/CAS9 system is a powerful tool for regulating damaged genome sequences. Nucleases that are damaged in their sequence are called miRNAs (micro RNAs). The miRNAs targeted by multiple promoter sgRNA (single guide RNA) are cut or regulated from RNA by the CRISPR/CAS9 method. The sgRNAs targeted to the incorrect miRNAs may provoke undesired genome abnormalities. In this study, in order to minimize these genome distortions, sgRNA target estimation was performed for CRISPR/CAS9 with deep learning in this study. In this article, convolutional neural networks (Convolutional Neural Networks-CNN) and multilayer perceptron (Multi- Layer Perceptron-MLP) algorithms are used for experimental analysis. We also compare the performance of CRISPR/CAS9 system for three algorithms.
Abbreviations: CNN: Convolutional Neural Networks; BLSTM: Bidirectional Long-Short Term Memory; MLP: Multi Layer Perceptron
Introduction
In an Esherichia Coli (E. Coli) bacteria that investigated by “In silico” (simulation) method which is important role nowadays has been discovered an immune system named CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 [1]. According to this system an E.Coli bacteria which is infected by any virus, add the virus DNA its memory and recognize when any other virus attack accrues. This defined virus DNA cuts this virus DNA from its own DNA through the CAS9 enzyme. Thus, DNA repair will occur. According to recent research the leading factor in gene regulation has been observed the “microRNA (miRNA)” targeted by the “single-guide RNA (sGRNA)” s. MiRNAs are small and noncoding RNA molecules [2]. In diseases, for example, different miRNAs involved in all stages of cancer make cancer diagnosis and treatment available [3]. CRISPR/CAS9 is used to destroy targeted miRNAs in cells [4]. It uses “guideRNA (gRNA)” as a guide to target the CRISPR/CAS9 nucleus to the DNA sequence and trigger the double-strand break at the desired location. Breaking and repair of these threads can cause random addition and alteration of DNA [5].
MiRNA activities are different in each cell type [6]. So, the miRNA activities of a human cell and a bacterial cell will not be the same. From this point, It is important to find a dataset about the genomes to be studied in this respect. Various miRNA target data sets used in “in silico” studies will be used [7]. The aim of this study, it is made mirRNA target estimation with machine learning algorithms. The result of wrong targeted miRNA may be cause of undesirable gene mutations [8]. It is aimed to minimize the Type 1 and Type 2 errors of miRNAs by applying machine learning and deep learning method of Type 1 and Type 2 errors as output. Thus, the wrong target estimate will be minimized. In this way, it will be possible to reliably repair gene damage by correctly targeting mistargeted miRNAs. Genetic diseases will be eliminated by repair of gene damage.
The purpose of the “in silico” technique is to increase the accuracy of disease prevention by single pointing with the help of mechanization. The large base readings provided by mechanization also contribute greatly. Studies for miRNA targeting, support vector machines (Support Vector Machine -SVM) [9], deep learning, constrained logic programming (Constraint Logic Programming) [10] and a class classification (One Class Classification-OCC) [11] methods were used. Besides, in the future big data may be decreased to its most valuable parts with digital data forgetting concepts. It makes computations faster and the machines will store less data in disks [37].
Materials and Methods
CRISPR
CRISPR/CAS9 system is a strong genome editing mechanism used in many biotechnology applications [12]. MiRNAs are RNA molecules that play an important role in gene regulation in animals and plants [13]. Damaged miRNAs are targeted by sgRNAs. However, the effectiveness of sgRNAs have not been defined firmly to target area [14]. In the systems of CRISPR/CAS9 (clustered regularly interspaced short palindromic repeats), Cas9 nuclease creates short array (about 20 nucleotide) with RNA guideline two stranded in the determined region at DNA. CRISPR/CAS9 has been a unique technology that allows genetic and medical researchers to add, subtract or modify DNA sequences in various parts of the genome. In contrast to ZFN (Zinc-Finger Nuclease) and TALENs (Transcription Activator-Like Effector Nuclease), CRISPR/CAS9 is not man-made; currently the system is part of the bacterial immune system, which helps to protect from invasive phages. Originality in CRISPR is achieved using an RNA molecule that is complementary to the gene of interest. After binding, this RNA molecule (also called guide RNA) traps a CAS9 nuclease, a double-strand break that causes a frame shift when repaired by NHEJ (Non-Homologous End Joining). CRISPR is the most effective process to date in gene repair and editing tools. Efficiency varies depending on the organism and the target site. Since it is the simplest, versatile and sensitive method of genetic manipulation that is currently available, it attracts great attention in the world of science. In summary, CRISPR/CAS9 differs from ZFN and TALENs in an important aspect that makes them superior to genomic regulation applications: The ZFN and TALENs bind to DNA through a direct protein-DNA interaction that requires redesigning the protein for each new target.
DATA Set
Applied “in silico” research and review, BLAST (Basic Local Alignment Search Tool) was used for CRISPR [15]. BLAST is a search tool that analyzes the amino acids and DNA sequences of proteins and finds similarities between them. Besides BLAST, the data set resources have been used such as National Human Genome Research Institute (NHGRI) [16], miRBase [17], Genome Crispr [18], CrisprInc [19], ENSEMBL [20], ENCODE [21], CRISPRz [22], CRISPOR [23], CRISPR Local [24]. In the algorithm studies performed with these data sets, estimation tools such as mirWalk [25], TargetScan (ID2 PPI analysis network) [26], miRanda [27], mirBase [28], mirTarget [29], TarBase [30] have been developed.
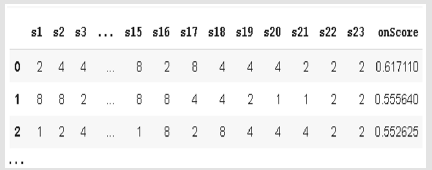
In this study, CRISPR Local data set has been used for the analysis [31]. The source of CRISPR Local dataset is ENSEMBL Plants. There are approximately 854.610 lines of CRISPR data in the original. In this study, we used 34.200 lines of data because of technical issues. There are 11 column features in this dataset which are examples of “Cyanidioschyzon merolae” alga. This features; the gene in which the sgRNA, on target estimated chromosome and its coordinate, sgRNA sequence with 23’nt., on target prediction score, off target prediction gene which has the greatest CFD score, the chromosome on target prediction, its coordinate and beginning position, off-target prediction sequence, the number of sgRNA and mismatch on the off-target sequence, axon name, axon start position, all off-target and sgRNA having the highest CFD score. The sequences having 4 channels like Adenin (A), Sitozin (C), Guanin (G) ve Timin (T) are used as [1,0,0,0], [0,1,0,0], [0,0,1,0], [0,0,0,11]. According these channels, the process has been realized with converted RNA sequence to binary system. These sequences were used as binary in the data set. In this example, each base in the sequence is considered as a separate column and feature. The 23nt. sgRNA sequence, on-target prediction score features were used. Figure 1 shows an illustration of the sample data set. used. Figure 1 shows an illustration of the sample data set.
Experimental Studies
In this study, CNN, MLP and BLSTM models were constituted and compared.
Multilayer Perceptron -MLP
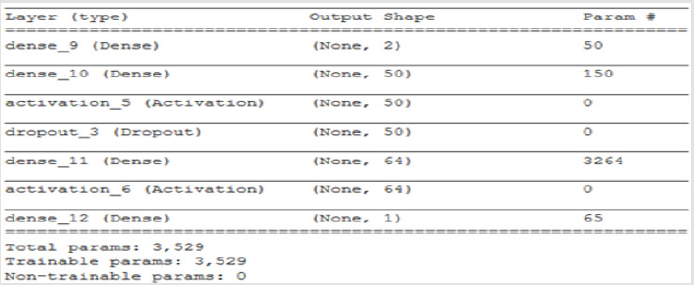
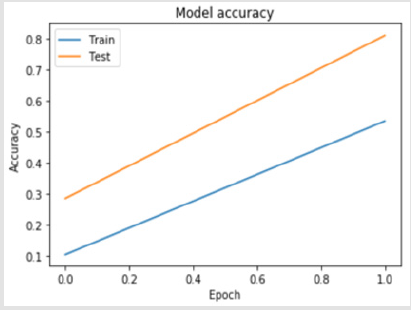
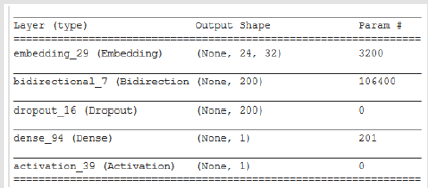
A 4-layer MLP model has been developed according to the MLP algorithm run using the Google Colab GPU (https://colab.research. google.com). In the MLP model a fully connected structure of dense layers was formed. Information on the model used is shown in Figure 2. The rates of logistic regression and accuracy according to the MLP model are shown in Figure 3 and Figure 4. Accuracy obtained 80.38% according to MLP model.
Convolutional Neural Network - CNN
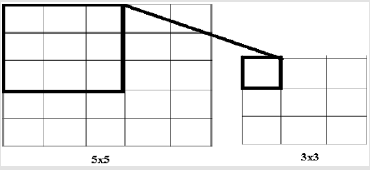
Convolutional Neural Networks are a model of artificial neutral network which is used successfully in image processing, bioinformatics, robotics, data mining, finance and many other areas. However, except for image analysis, surprisingly high accuracy ratio was obtained in emotion analysis, text classification and question answering applications. According to this model, it is applied to nxn matrix with nxn filtering method (dot product), with acceptation of n>m. Thus, it allows the identification and classification of properties. In Figure 5, 3x3 matrix as a result of the intrinsic product of a 5x5 matrix and filtering was obtained. In this study, data set has been divided two group one of training 70 per cent, the other test 30 per cent. The model has been fixed up to non-linear by using the tangent and sigmoid activation functions. Convolution network is used to clarify the properties. The convolution network helps to create a new matrix with the results of the multiplication of the matrices. In order to prevent over fitting, maxpooling layer was used. It selects the elements with the maximum value from the matrix pool of the specified size in the maxpooling layer. Accordingly, the information obtained when the CNN model is generated can be seen in Figure 6. The rate of accuracy according to the CNN model can be seen from Figure 7.
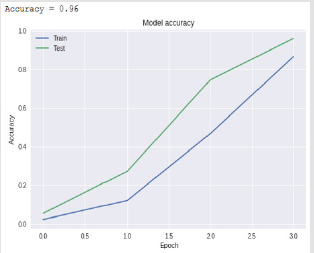
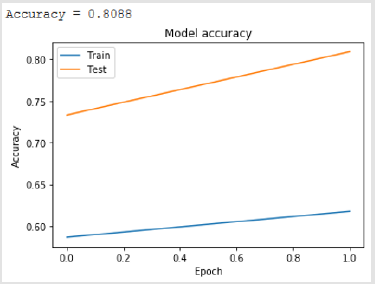
Bidirectional Long-Short Term Memory - BSLTM
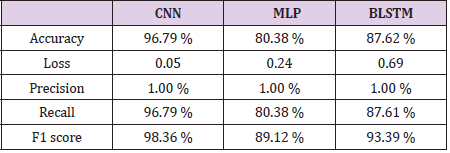
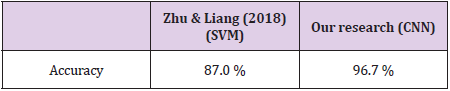
Bidirectional LSTM is different from other feed forward models in neural networks feedback system. Accordingly, the information obtained when the bidirectional LSTM model is generated can be seen in Figure 8. The rate of accuracy according to the bidirectional LSTM model can be seen from Figure 9. Accuracy has obtained 80.88 % with bidirectional LSTM model. Accuracy was 96.7% compared to the CNN model. Stochastic Gradient Descent (SGD) optimization method was used. Learning rate was set to 0.0005. Binary cross entropy logarithmic regression function was used. The following table (Table 1) shows the MLP, CNN and bidirectional LSTM model accuracy rates. Table 2 shows the MLP, CNN and bidirectional LSTM model accuracy rates, precision-recall, f-measure values. In another study [35] authors used the same data set. Source of Crispr Local which is Ensembl Plants [36] data contains on a set of plant and animal cell data. Support Vector Machine (SVM) algorithm was applied. Located accuracy of ratio is 87 %. Table 3 shows a comparison of our algorithm and last studies accuracy rate.
Conclusion
In this study, the algorithms of Multilayer Perceptron-MLP, Convolutional Neural Networks-CNN and Bidirectional Long Short- Term Memory-BLSTM have been compared with use of CRISPR data set. As a result, according to this data set, the accuracy rate in the MLP model was 80.38 % and Bidirectional LSTM model was 80.88 % whereas for CNN this result was found 96.7 %. According to the results, a highest accuracy rate was obtained with the CNN model than MLP and BLSTM. In the CNN model, revised CRISPR has reached up to 7 layers according to the ENSEMBL Plants dataset and 4 layers have been formed in MLP and 2 layers have been formed in BSLTM. Comparing other algorithms with our algorithm is better performance according to results. Research that used with SVM algorithm performed 87.0 % accuracy result. However, our model achieved 96.7 %. This result is more reliable performance than the research used SVM algorithm. Any mistargeted position causes unwanted genome distortions. For this reason, the accuracy rate is urgent in sgRNA targeting.
References
- R Wilkinson, B Wiedenheft (2014) A CRISPR method for genome engineering. F1000prime reports 6(3).
- H Chang, Bin Yi, Ruixia Ma, Xiaoguo Zhang, Hongyou Zhao, et al. (2016) CRISPR/CAS9, a novel genomic tool to knock down microRNA in vitro and in vivo. Scientific reports 6: 22312.
- G Aquino Jarquin (2017) Emerging role of CRISPR/Cas9 technology for MicroRNAs editing in cancer research. Cancer Research 77(24).
- JS Kurata, RJ Lin (2018) MicroRNA-focused CRISPR-Cas9 library screen reveals fitness-associated miRNAs. RNA 24(7): 966-981.
- A Pla, Xiangfu Zhong, Simon Rayner (2018) miRAW: A deep learning-based approach to predict microRNA targets by analyzing whole microRNA transcripts. PloS Comput Biology 14(7).
- M Sualp, T Can (2011) Using network context as a filter for miRNA target prediction. Biosystems 105(3): 201-209.
- A John, Angela Schoolmeesters, Eldon T Chou, Elena Maksimova, et al. (2017) Using the CRISPR-Cas9 system with paired Dharmacon™ Edit-R™ synthetic crRNAs for functional knockout of microRNA hsa-miR-221. Dharmacon, A Horizon Discovery Group Company, USA.
- M Hirosawa, Fujita Y, Parr CJC, Hayashi K, Kashida S, et al. (2017) Cell-type-specific genome editing with a microRNA-responsive CRISPR-Cas9 switch. Nucleic Acids Research 45(13): e118.
- X H Zhang, Tee LY, Wang XG, Huang QS, Yang SH (2015) Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Molecular Therapy-Nucleic Acids 4: e2643.
- M T Tekbulut (2006) MicroRNA target prediction by constraint programming Thesis (Master). Sabancı University, Istanbul.
- M Sualp (2013) Machine Learning Methods for Using Network Based Information in Microrna Target Prediction. Middle East Technical University Computer Engineering.
- DP Bartel (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2): 215-233.
- S Abadi (2017) A machine learning approach for predicting CRISPR-Cas9 cleavage efficiencies and patterns underlying its mechanism of action. PLOS Computational Biology 13: 10.
- F Stephen Altschul, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17): 3389-3402.
- D Welter, MacArthur J, Morales J, Burdett T, Hall P, et al. (2014) The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Research 42(Database issue): D1001-D1006.
- S Griffiths, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36(Database issue): D154-D158.
- B Rauscher, Florian Heigwer, Marco Breinig, Jan Winter, Michael Boutroset (2017) GenomeCRISPR - a database for high-throughput CRISPR/Cas9 screens. Nucleic Acids Research 45(Database issue):D679–D686.
- J Cohen (2017) The Birth of CRISPR Inc. Science 355.6326: 680-684.
- A Yates, Wasiu Akanni, M Ridwan Amode, Daniel Barrell, Konstantinos Billis et al., (2016) Ensembl 2016. Nucleic Acids Research 44(Database issue): D710-D716.
- EA Feingold (2004) The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306(5696): 636-640.
- G K Varshney, Suiyuan Zhang, Wuhong Pei, Ashrifia Adomako Ankomah, Jacob Fohtung, et al. (2016) CRISPRz: a database of zebrafish validated sgRNAs. Nucleic Acids Research 44(Database issue): D822-D826.
- M Haeussler, Schönig K, Eckert H, Eschstruth A, Mianné J, et al. (2016) Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biology 17(1):148.
- J Sun, Liu H, Liu J, Cheng , Peng Y, et al. (2018) CRISPR-Local: a local single-guide RNA (sgRNA) design tool for non-reference plant genomes. Bioinformatics 35(14): 2501-2503.
- H Dweep, Sticht C, Pandey P, Gretz (2011) miRWalk – Database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. Journal of Biomedical Informatics 44(5): 839-847.
- Y Shi, Yang F, Wei S, Xu G (2017) Identification of Key Genes Affecting Results of Hyperthermia in Osteosarcoma Based on Integrative ChIP-Seq/TargetScan Analysis. Med Sci Monit 23: 2042-2048.
- E AJ, John B, Gaul U, Tuschl T, Sander, et al. (2003) MicroRNA targets in Drosophila, Genome Biology 5(1): R1.
- S Griffiths Jones, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Research 36(Database issue): D154-D158.
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, et al. (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7): e47.
- P Sethupathy, Corda B, Hatzigeorgiou AG. (2006) TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA 12(2): 192-197.
- H Liu, Liu H, Liu J, Cheng S, Peng Y, et al. (2018) CRISPR-Local: a local single-guide RNA (sgRNA) design tool for nonreference plant genomes. Bioinformatics 35(14): 2501-2503.
- L Jiecong, W Ka Chun (2018) Off-target predictions in CRISPR-Cas9 gene editing using deep learning ECCB 2018 Proceeding Special Issue. Bioinformatics 34(17): 656-663.
- C Guohui, Ma H, Yan J, Chen M, Hong N, et al. (2018) Deep CRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biology 19(1): 80.
- J Listgarten, Weinstein M, Kleinstiver BP, Sousa AA, Joung JK, et al. (2018) Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs. Nat Biomed Eng 2(1): 38-47.
- H Zhu, C Liang (2019) CRISPR-DT: designing gRNAs for the CRISPR-Cpf1 system with improved target efficiency and specificity. Bioinformatics 35(16): 2783-2789.
- D Bolser, Staines DM, Pritchard E, Kersey P (2016) Ensembl Plants: Integrating Tools for Visualizing, Mining, and Analyzing Plant Genomics Data. Methods Mol Biology 1374: 115-140.
- M Gunay, E Yildiz, Y Nalcakan, B Asiroglu, A Zencirli, et al. (2018) Digital Data Forgetting: A Machine Learning Approach. IEEE International Symposium on Multidisciplinary Studies and Innovative Technologies p. 1-4.

 Research Article
Research Article